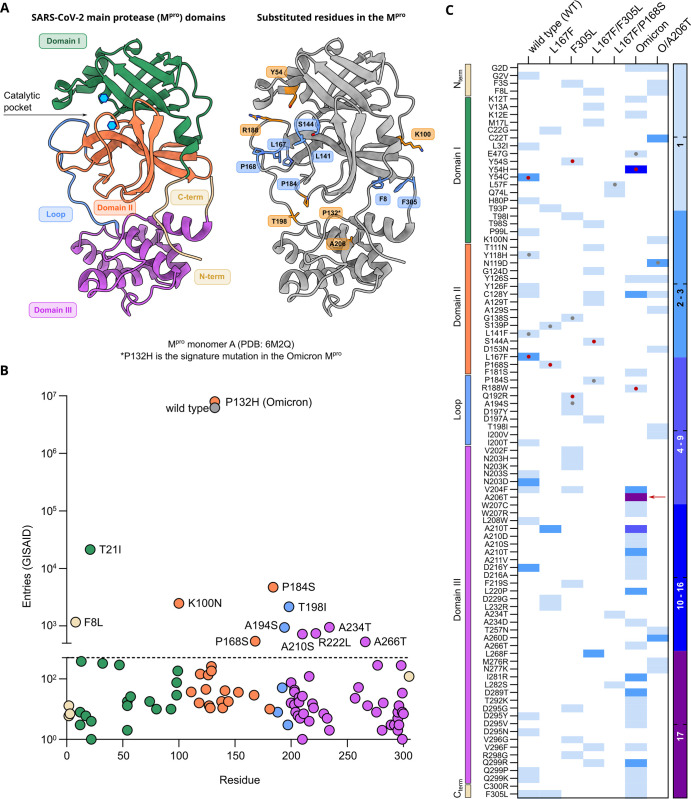
Fig 2. Mpro structure, Frequency of Mpro mutations in the GISAID database and generated mutations.
(A) 3D ribbon structure of the Mpro monomer A (PDB: 6M2Q [44]). On the left side of the panel, Mpro is colored according to described domains [42]. Catalytic dyad (H41 and C145) is indicated by the light blue hexagons. On the right side of the panel, the same Mpro structure is shown highlighting some of the residue positions that were found to be mutated after selection experiments: residues found mutated in the WT background are colored blue; mutants in the Omicron background are colored in orange. Mutants at these residues were investigated either alone or in specific combinations. (B) The total number of nsp5/Mpro GISAID substitution entries for each mutation found after selection experiments. Entries are colored according to domains as displayed in panel A. WT and Omicron-Mpro sequences are displayed in grey and orange, respectively. The dotted line at 500 represent the cut-off value. (C) Heat map representing the number of a specific substitution’s independent occurrences from different samples of VSV-Mpro selection experiments. Wild type and F305L columns comprise mutants that were selected in previous work [35], as also shown in Fig 1D. Color-coded domains are indicated as in A. The red arrow indicates the substitution A206T. Red dots indicate residues within the catalytic site and grey dots indicate residues near the catalytic site.