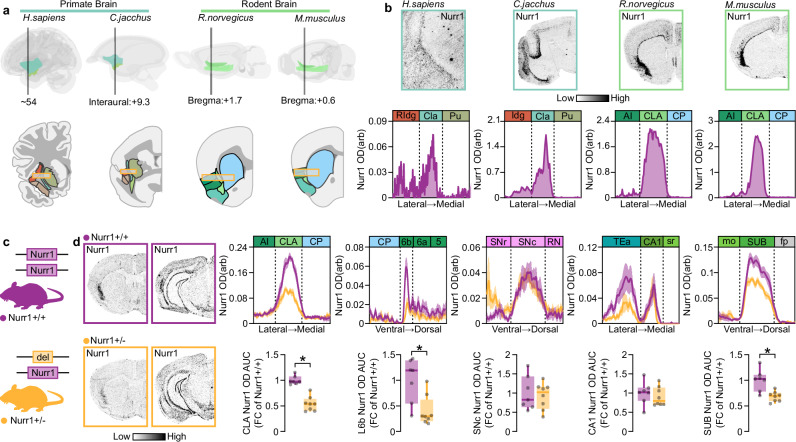
Fig. 1. CLA Nurr1 mRNA is conserved among species and is dependent on Nurr1 gene dosage.
a Schematic depiction of the CLA in primate (H. sapiens and C. jacchus) and rodent (R. norvegicus and M. musculus) brain and its location between the Insular cortex and Striatum. Yellow framed rectangles are centered on CLA and delineate the axis across the average OD traces were measured b Nurr1 mRNA is enriched in human (H. sapiens), marmoset (C. jacchus), rat (R. norvegicus) and murine (M. musculus) CLA. c Illustration of Nurr1+/− mouse line. d Left: ISH autoradiographs showing Nurr1 mRNA at CLA and SNc level. Right: Line-graphs (upper) and boxplots (lower) showing the Nurr1 mRNA OD quantification in CLA, L6b, SNc, CA1, and SUB of Nurr1+/+ and Nurr1+/− mice (Nurr1+/+: n = 7, Nurr1+/−: n = 8; CLA: *p < 0.0001, L6b: *p = 0.0067, SUB: *p = 0.0021, Two-sided Unpaired t-test). Data in line-graphs are expressed as mean ± SEM. Boxplots show all data points, the 25th and 75th percentile (box), the median (center), and the maxima (whiskers). OD: optic density, arb: arbitrary units, RIdg: rostral dysgranular insular cortex, Idg: insular dysgranular cortex, Cla: human claustrum/dorsal endopiriform cortex complex/dorsal endopiriform cortex complex, Pu: putamen, AI: agranular insula, CLA: rodent claustrum/dorsal endopiriform cortex complex/dorsal endopiriform cortex complex, CP: caudoputamen, AUC: area under the curve, FC: fold change, L6b: cortical layer 6b, SNr: substantia nigra reticular part, SNc: substantia nigra compact part, RN: reticular nucleus, TEa: temporal association areas, sr: stratum radiatum of CA1, mo: molecular layer of dentate gyrus, SUB: subiculum, fp: posterior forceps of corpus callosum. ISH: in situ hybridization. Source data are provided as a Source Data file.