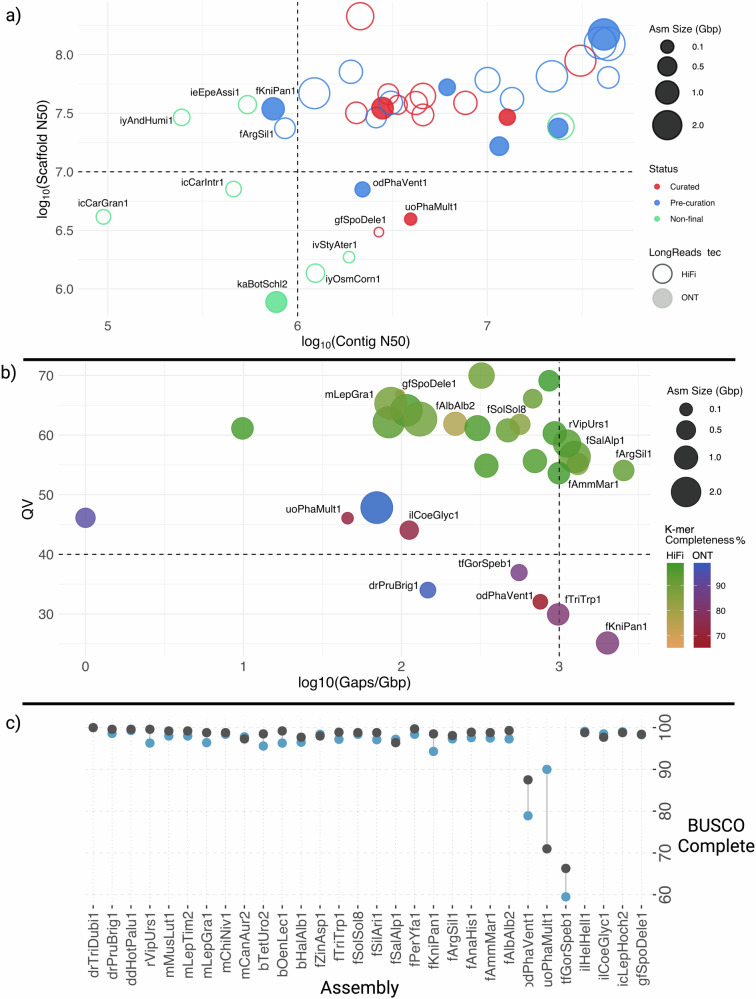
Fig. 4. Quality control and status of the 38 genome assemblies evaluated.
a Genome assemblies are represented according to their Scaffold N50 (y-axis, log10) and number of the longest scaffolds that comprise at least 95% of the assembly (x-axis, log2). Bubble size is proportional to assembly span. Empty bubbles depict HiFi-based genomes, while full bubbles are ONT-based. Colours are according to assembly status (Curated, Pre-curation, Non-final draft). Lower values for both axes indicate better assembly contiguity. Assemblies not reaching the EBP-recommended One Megabase Contig N50 (log101,000,000 = 6) or 10 Megabase Scaffold N50 (log1010,000,000 = 7) here a proxy for chromosome-level scaffolds are labelled with their ToLIDs* (https://id.tol.sanger.ac.uk/). b Completed HiFi- and ONT-based genomes assemblies are represented according to their Quality value (QV, y-axis) and number of gaps per Gbp (log10, x-axis). The bubble size is proportional to assembly size. Colour grade of the bubbles is according to the K-mer completeness score. ToLIDs are reported for the assemblies that are below the recommended EBP metric for QV (40), Gaps/Gbp (log101000 = 3) or K-mer completeness (90%). Quality values are calculated differently for HiFi-based assemblies than for ONT-based assemblies and should not be compared directly. c BUSCO completeness scores for genome assemblies with ‘Curated’ and ‘Pre-curation’ status. Using two orthologs databases, one for a more recent last common ancestor encompassing related species (blue), and one for all eukaryotes (grey), we seek a more comprehensive estimation of the assembly completeness. Number of single-copy orthologs present on each database is reported. *Briefly, a ToLID is a unique identifier for an individual organism within a species sampled for genome sequencing, consisting of one or two lowercase letters for high-level taxonomic rank and clade, respectively, followed by three letters for genus and species each. Thus, within insects (i), the Hemiptera (i) includes Andrena humilis (iyAndHumi1) and Osmia cornuta (iyOsmCorn1). The Coleoptera (c) contains Carabus granulatus (icCarGran1), C. intricatus (icCarIntr1), and Leptodirus hochenwarti (icLepHoch2). Ephemeroptera (e) features Epeorus assimilis (ieEpeAssi1), and among Strepsiptera (v) it is found Stylops ater (ivStyAter1). Lepidoptera (l) includes Coenonympha glycerion (ilCoeGlyc1), Helleia helle (ilHelHell1), and Parnassius mnemosyne (ilParMnem1). Within the fungi (g), Agaricomycetes (f) are represented by Spongipellis delectans (gfSpoDele1). For sponges (o), Demospongiae (d) includes Phakellia ventilabrum (odPhaVent1), and among algae (u), Heterokontophyta (o) are represented by Phaeosaccion multiseriatum (uoPhaMult1). The fishes (f) include Alburnus alburnus (fAlbAlb2), Ammodytes marinus (fAmmMar1), Anaecypris hispanica (fAnaHis1), Argentina silus (fArgSil1), Knipowitschia panizzae (fKniPan1), Perca sp.‘yellow fin Alpine’ (fPerYfa1), Salvelinus alpinus (fSalAlp1), Silurus aristotelis (fSilAri1), Solea solea (fSolSol8), Tripterygion tripteronotum (fTriTrp1), and Zingel asper (fZinAsp1). Birds (b) are represented by Haliaeetus albicilla (bHalAlb1), Oenanthe leucura (bOenLec1), and Tetrao urogallus (bTetUro2). Mammals (m) include Canis aureus (mCanAur2), Chionomys nivalis (mChiNiv1), Lepus granatensis (mLepGra1), Lepus europaeus (mLepEur2), and Mustela lutreola (mMusLut1). Among reptiles (r) is Vipera ursinii (rVipUrs1). Within dicotyledons (d), the Ericales (d) include Hottonia palustris (ddHotPalu1), and Rosales and Fabales (r) features Prunus brigantina (drPruBrig1) and Trifolium dubium (drTriDubi1), respectively. Finally, among ‘other chordates’ (k), Ascidiacea (a) includes Botryllus schlosseri (kaBotSchl2), while in the category ‘other animal phyla’ (t), Nematomorpha (f) is exemplified by Gordionus montsenyensis (tfGorSpeb1).