FIGURE 1.
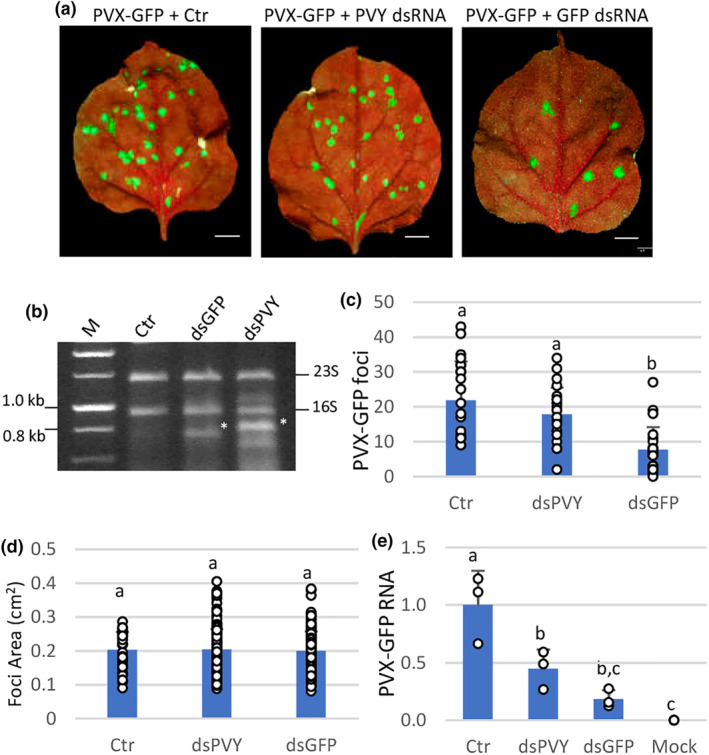
Sequence‐specific and nonspecific dsRNAs interfere differently with PVX‐GFP local infection. Nicotiana benthamiana plants were inoculated with mixtures of PVX‐GFP combined with nucleic acid extracts prepared from Escherichia coli accumulating the dsPVY or dsGFP. (a) Representative inoculated leaves were examined under UV light at 7 days post‐inoculation (dpi). Scale bar denotes 1 cm. (b) E. coli HT115(DE3) cultures transformed with L4440 encoding either a dsRNA consisting of 902 bp of the PVY coat protein (CP) gene (dsPVY) or GFP (dsGFP) were induced with IPTG and processed for total nucleic acid. The total nucleic acid extracted from bacterial cells not expressing dsRNA was used as a control (Crt). Samples were resolved by electrophoresis on 1% agarose gel. The positions of 23S and 16S rRNA are indicated on the right. dsRNA bands are indicated by asterisks. NZYDNA Ladder III was used as dsDNA markers (M). (c) Mean numbers ± SD of infection foci on inoculated leaves of 12 plants (two leaves per plant) with the different treatments at 7 dpi. (d) Mean sizes ± SD of infection foci on inoculated leaves of six plants with the different treatments. (e) Reverse transcription‐quantitative PCR was used to analyse the accumulation of PVX‐GFP genomic RNA levels in the inoculated leaves at 7 dpi. Mock, mock‐inoculated plants. Expression of the 18S rRNA gene served as a control. Data represent the means ± SD of three replicates, each consisting of a pool of 12 plants that received the same treatment. Different letters indicate significant differences determined by employing Scheffé's multiple range test (p < 0.05). Experiments were repeated once more with similar results.
