FIGURE 3.
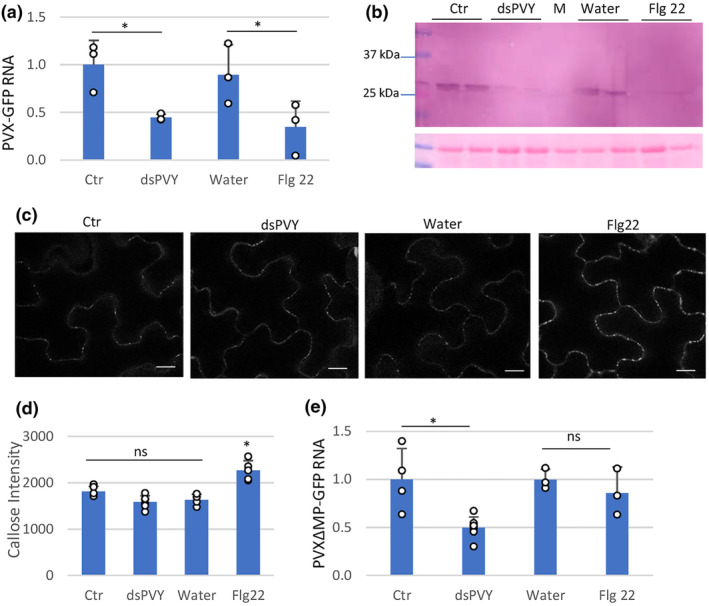
Nonspecific dsRNA elicits inhibition of PVX‐GFP replication. Plants were inoculated with mixtures of PVX‐GFP and either flagellin (flg22), dsPVY, control (Ctr) extracts or water. (a) Reverse transcription‐quantitative PCR (RT‐qPCR) was used to analyse the accumulation of PVX‐GFP genomic RNA levels in the inoculated leaves at 4 days post‐inoculation (dpi). Expression of the 18S rRNA gene served as a control. Data represent the means ± SD of three replicates, each consisting of a pool of nine plants that received the same treatment. (b) Western blot analysis of plant extracts derived from inoculated leaves at 4 dpi, using antibodies against PVX CP. The panel below the blot is the membrane stained with Ponceau S as control of loading. M, mock‐inoculated leaf. (c) Callose spots at plasmodesmata (PD) were visualized upon aniline blue staining of epidermal cells in response to water, bacterial flg22, dsPVY or Ctr extracts. Photographs were taken 30 min after treatment with water, 1 μM flg22, 30 ng/μL of dsPVY or Ctr extracts. Scale bar, 10 μm. (d) Relative PD callose content in water‐, flg22‐, dsPVY‐ and Ctr‐treated leaves. The mean values of callose intensities in individual PD (>200) measured in three leaf discs taken from two independent biological replicates per each treatment. (e) Accumulation of a frameshift mutant in the P25 movement protein of PVX‐GFP (PVXΔMP‐GFP), as assayed by RT‐qPCR at 4 dpi. Asterisks indicate significant differences between treatments (Student's t test, *p < 0.05); ns, not significant. Experiments were repeated once more with similar results.
