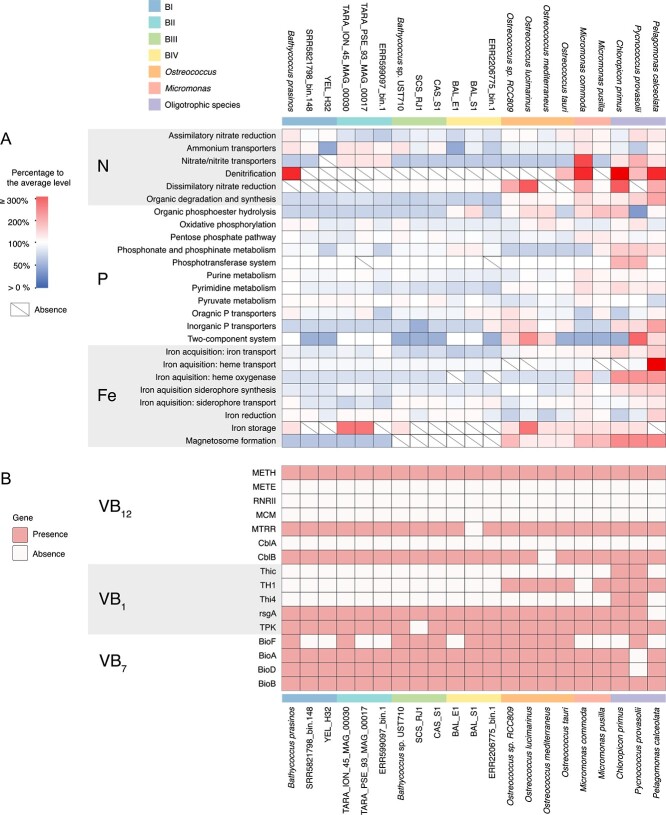
Figure 4.
Comparison of nutrient metabolism gene content among eukaryotic picophytoplankton. The selected 21 genomes of eukaryotic picophytoplankton include four Bathycoccus clades, Micromonas, Ostreococcus, and three oligotrophic species. (A) The heatmap depicts differences in gene content involved in nitrogen (N), phosphorus (P), iron (Fe) metabolism among the eukaryotic picophytoplankton. The heatmap gradient indicates whether the gene copy number for a specific process is overrepresented, equally represented, or underrepresented compared with the average level of the selected genomes. Boxes with a diagonal line indicate the absence of genes associated with a particular process. (B) The binary heatmap displays the presence (red) or absence (white) of genes encoding vitamin B12 (VB12)-dependent enzymes (METH, RNRII, MCM), VB12-independent enzyme (METE), and their accessory proteins (MTRR, CblA, CblB), as well as proteins involved in biosynthesis of vitamin B1 (VB1) and vitamin B7 (VB7).