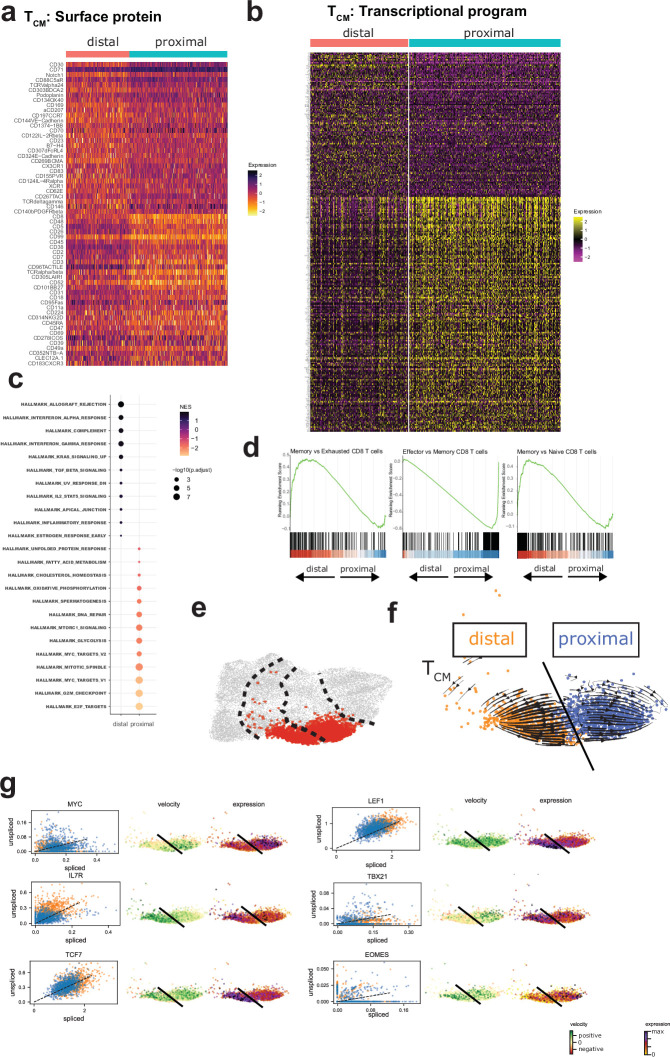
Extended Data Fig. 8. TCM-like proximal-daughter and distal-daughter CD8 CARTs exhibit asymmetry in surface proteomic and transcriptional landscapes that drive effector or memory differentiation programs.
a-h, Surface proteomic and transcriptional profile asymmetry in first-division TCM-like daughter CARTs support memory maintenance in distal cells and proliferative and effector differentiation in proximal cells. (a) Heat map of normalized surface protein levels of the top 30 proteins enriched in either distal or proximal cells. (b) Heat map of normalized gene expression of top enriched genes in distal-daughters (180 genes) and proximal-daughters (240 genes). (c) Hallmark transcriptional programs of distal and proximal TCM-like CARTs support increased metabolic activity (fatty acid metabolism, oxidative phosphorylation, glycolysis, MTORC1 signaling) and proliferation (MYC targets, mitotic spindle, G2M checkpoint, E2F targets) in proximal-daughters compared to distal-daughters. Statistical significance was determined using GSEA test with Benjamini–Hochberg correction for multiple comparisons. (d) Gene-set enrichment plots comparing transcriptional programs between distal-daughter and proximal-daughter TCM-like CARTs demonstrating enrichment in memory-associated programs in distal cells and effector-associated programs in proximal cells. (e) Proximal and distal TCM clusters (clusters 11 and 5) characterized in f-g. (f) Velocity vector projection onto TCM UMAP clusters with streamline plots indicating divergent cell-state transitions between proximal and distal daughter cells. Black line signifies border between distal (orange) and proximal (blue) cells. (g) Gene-specific RNA velocity displayed as spliced/unspliced transcripts (left column) and projected onto TCM UMAP clusters (middle column), with normalized gene expression levels as a comparison (right column), demonstrate that both intrinsic transcriptional changes (MYC upregulation in proximal-daughters, IL7R upregulation in distal-daughters) and asymmetric assortment of pre-existing RNA (greater abundance of TCF7, LEF1 in distal-daughters with similar RNA velocities as proximal-daughters) are mechanisms for transcript abundance differences during ATCD. Plots are representative of 2 independent experiments with distinct donors: one with the anti-TCRδ CAR (shown in this figure) and one with the anti-CD19 CAR.