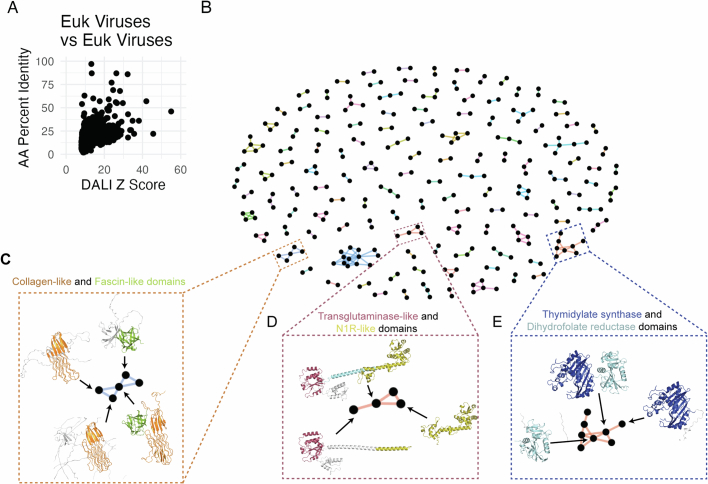
Extended Data Fig. 6. Shared domains across eukaryotic virus protein clusters.
A. All-by-all structural alignments of representative structures from the 5,770 protein clusters with more than one member. Each dot indicates a single alignment, with the Y axis indicating the fraction of amino acid identity and the X axis indicating DALI Z-score. B. Protein clusters tend to share protein domains. Each node indicates a protein cluster, and edges between protein clusters indicate there is a DALI alignment between them. Only alignments with a Z score of at least 15 are plotted. The boxes indicate cluster representatives highlighted in subsequent panels. C. Frequent reuse of structural/cytoskeleton-related domains. Protein clusters with collagen-like domains (orange) and fascin-like domains (green) are highlighted. D. Multiple combinations of domains with the same viral genus. Diverse combinations of transglutaminase-like domains (purple) and N1R-like domains (green) from entomopoxvirus proteins are highlighted. E. Frequent reuse of protein domains involved in metabolism. Various combinations of thymidylate synthase (dark blue) and dihydrofolate reductase (light blue) domains in protein clusters are highlighted.