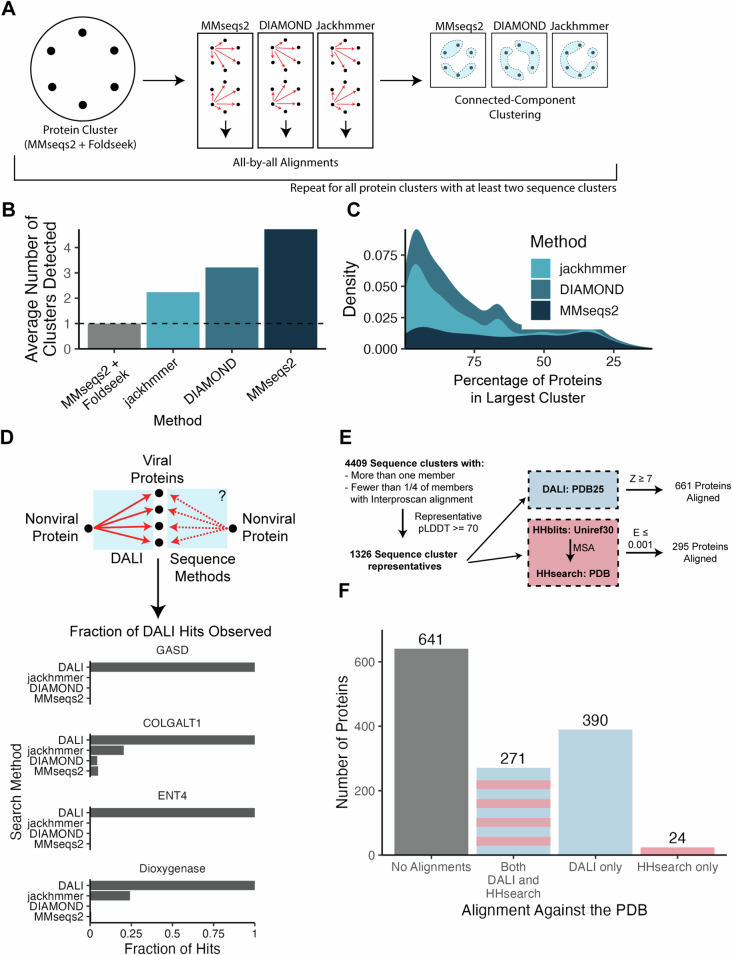
Extended Data Fig. 7. Structure methods outperform sequence methods at identifying virus-virus and host-virus protein similarities.
A. Method for doing benchmarking. For all protein clusters with at least two sequence clusters, we conducted all-by-all alignments between members using MMseqs2, DIAMOND blastp, and jackhmmer. These alignments and subsequence clustering occur separately for each protein cluster. From these alignments, we conducted connected-component clustering using sat.py aln_cluster. B. This plot indicates the average number of clusters detected (on the Y axis) by each method (on the X axis) across all of the protein clusters that contain at least two sequence clusters. C. For each sequence method, the proportion of original protein cluster members that were included in the largest cluster is plotted across all original protein clusters. The X axis indicates the proportion of proteins in the largest cluster for the indicated sequence method (color), while the Y axis indicates the density (or proportion) of original protein clusters with that value. D. To compare the sensitivity of structure and sequence alignment at detecting similarities between virus and non-virus proteins, we conducted sequence alignments using MMseqs2, DIAMOND, and jackhmmer to align each non-viral query against the viral database. These plots then indicate, for each query, the fraction of DALI alignments that are likewise identified through each sequence method. E. We identified swell-folded sequence cluster representatives from clusters containing no more than ¼ of members with an Interproscan alignment. We aligned these 1,326 proteins against the PDB using structural search (with DALI) or sequence search (with HHblits and HHsearch, similar to HHpred webserver). This resulted in 661 alignments with DALI and 295 alignments with HHblits/HHsearch. F. This bar plot indicates, for each of the 1,326 proteins in the benchmark set, the number of proteins with no alignments against the PDB with either DALI or HHblits/HHsearch, alignments against the PDB with both DALI and HHblits/HHsearch, or alignment against the PDB with only DALI or HHblits/HHsearch.