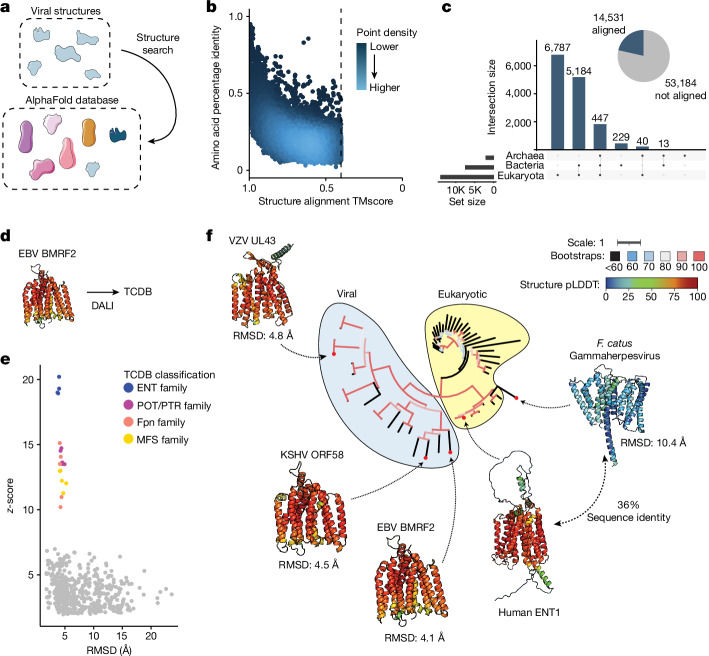
Fig. 3. Structural similarity across kingdoms of life reveals potential protein function.
a, Illustration of the approach. The database of viral protein predicted structures was aligned against the AlphaFold database of proteins from 48 organisms, including members of the bacterial, eukaryote and archaeal superkingdoms. b, The amino acid percentage identity and Foldeseek TMscore; each point indicates a single alignment. For viral proteins with more than five alignments, the top five alignments by TMscore are plotted. c, Right, pie chart indicating the number of viral proteins that do or do not have an alignment against the AlphaFold database. Left, UpSet plot indicating, for those viral proteins with alignments against the AlphaFold database, the number that align against members of each superkingdom. d, EBV BMRF2 (YP_001129455), which has a nucleoside transporter-like fold, was used as a query for a DALI search against the TCDB. e, Alignments between EBV BMRF2 and structures classified in the TCDB. Each dot indicates a single DALI alignment. Proteins with at least one alignment with z ≥ 10 are coloured. RMSD, root mean squared deviation as determined by DALI. f, A phylogenetic tree of eukaryotic and herpesvirus nucleoside transporters. The listed RMSD values were determined by DALI alignment between human ETN1 and each viral nucleoside transporter. The tree scale is substitutions per residue. Structures are coloured by pLDDT (red, higher; blue, lower). The tree is coloured according to bootstrap values. Accessions: F. Catus gammaherpesvirus, YP_009173937; VZV UL43, NP_040138; EBV BMRF2, YP_001129455; KSHV ORF58, YP_001129415; human ENT1, XP_011512643.