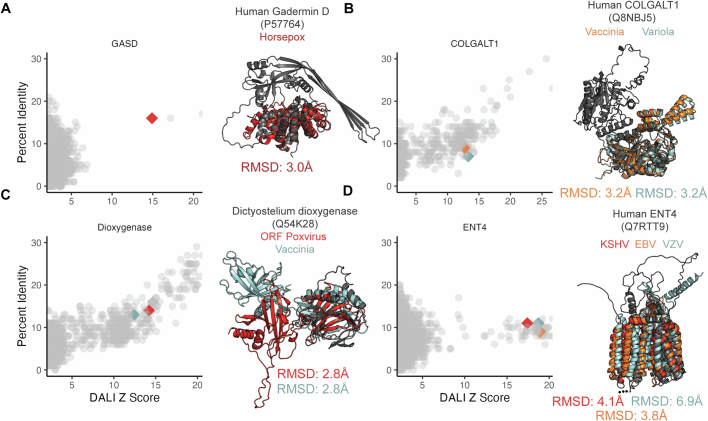
Extended Data Fig. 4. Structural similarities between viral and non-viral proteins.
A-D. Specific non-viral hits from the Alphafold Foldseek search were aligned against the viral predicted structure database using DaliLite, and alignments against proteins from human pathogens were selected. (Left) The Y axis indicates the percentage amino acid identity, and the X axis indicates the Dali Z score. Each dot indicates a single alignment. Each point indicates an alignment, with the points corresponding to the proteins highlighted on the right as diamonds and colored consistently with their protein structures. (Right) The structure of the non-viral protein query is present in black. A superposition of selected protein clusters is shown, with the RMSD of each viral protein vs the non-viral protein indicated. Protein accessions are as follows: GASD: (Horsepox-ABH08278). COLGALT1: (Vaccinia-YP_232983; Variola-NP_042130). Dioxygenase: (ORF Poxvirus-NP_957891; Vaccinia-YP_232906). ENT4: (KSHV-YP_001129415; VZV-NP_040138; EBV-YP_401658).