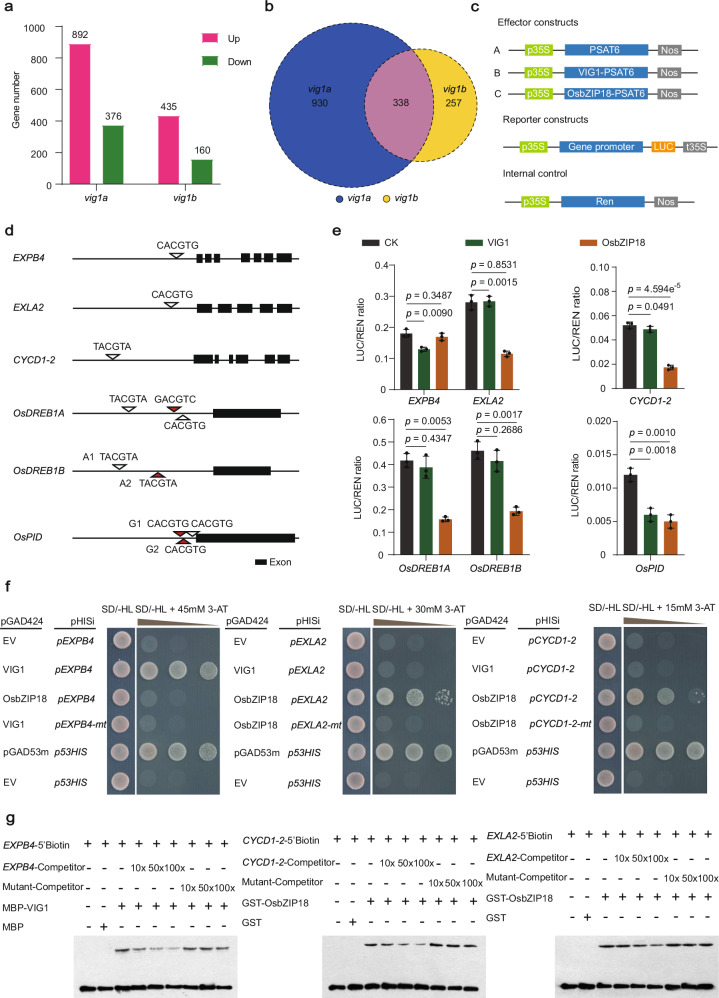
Fig. 5. VIG1 functions with OsbZIP18 in repressing downstream genes connected with cell expansion, cell division, chilling tolerance, and grain number per panicle.
a A total of 1268 and 595 DEGs were identified in vig1a and vig1b, respectively. b A total of 338 DEGs (indicated by the pink color) were found co-regulated in both vig1a and vig1b. c–g Analysis of the binding of VIG1 and OsbZIP18 to target gene promoters using the luciferase reporter (LUC) system, yeast one-hybrid (Y1H), and electrophoretic mobility shift assays (EMSA). c Schematic diagram of the constructs used for transactivation activity assay. d Scanning of the VIG1 binding motif ‘C/T/GACGTG/A/C’ across the promoter region of genes downregulated in vig1a. The length of promoter sequence is 2 kb for each gene. e Transcriptional repression of VIG1 and OsbZIP18 on downstream targets. Values are the mean ± SD (n = 3 biological replicates, two-tailed t-tests). f, g VIG1 directly binds to the promoter of EXPB4, while OsbZIP18 binds to the promoter of EXLA2 and CYCD1-2 which are confirmed by Y1H (f) and EMSA (g). The pGAD53m-p53HIS and pGAD424-p53HIS combinations are taken as the positive and negative controls, respectively, in the Y1H assays. The experiments in (f, g) are repeated at least three times with similar results. Source data are provided as a Source Data file.