Abstract
Clostridioides difficile infection (CDI) is a common healthcare-associated infection and the leading cause of gastroenteritis-related deaths worldwide. To investigate the effects of peptide composition of different protein products on CDI, we analyzed and compared the peptide sequences and compositions from Engraulis japonicus and Glycine max using Ultra High Performance Liquid Chromatography Tandem Mass Spectrometry (UPLC-MS/MS). An animal model of CDI was also established to investigate the potential therapeutic effects of these peptides in vivo. The peptide compositions of E. japonicus and G. max differed, with only 11% of the peptide sequences being identical. Oral administration of the tested peptides could reduce intestinal inflammation, repair the intestinal barrier, increase the proportion of beneficial bacteria, and reduce the proportion of harmful bacteria, providing a therapeutic effect against CDI. However, the peptides may differ considerably in some aspects. E. japonicus peptides were superior to G. max peptides in promoting colon epithelial cell proliferation and repairing tight intestinal cell junctions. Interestingly, the two sources of peptides have different effects on the cecal microbiome. E. japonicus peptides can effectively restore the diversity and richness of intestinal microbiota, while G. max peptides have poor regulatory effects on the intestinal microbiota structure. Overall, E. japonicus peptides showed better results than G. max peptides in treating CDI. This study supports the potential treatment of CDI with natural peptides and promotes the development of specialty foods for CDI enteritis. Clostridioides difficile infection (CDI) is a common healthcare-associated infection and the leading cause of gastroenteritis-related deaths worldwide. To investigate the effects of peptide composition of different protein products on CDI, we analyzed and compared the peptide sequences and compositions from Engraulis japonicus and Glycine max using Ultra High Performance Liquid Chromatography Tandem Mass Spectrometry (UPLC-MS/MS). An animal model of CDI was also established to investigate the potential therapeutic effects of these peptides in vivo. The peptide compositions of E. japonicus and G. max differed, with only 11% of the peptide sequences being identical. Oral administration of the tested peptides could reduce intestinal inflammation, repair the intestinal barrier, increase the proportion of beneficial bacteria, and reduce the proportion of harmful bacteria, providing a therapeutic effect against CDI. However, the peptides may differ considerably in some aspects. E. japonicus peptides were superior to G. max peptides in promoting colon epithelial cell proliferation and repairing tight intestinal cell junctions. Interestingly, the two sources of peptides have different effects on the cecal microbiome. E. japonicus peptides can effectively restore the diversity and richness of intestinal microbiota, while G. max peptides have poor regulatory effects on the intestinal microbiota structure.
Subject terms: Bacterial toxins, Bacterial pathogenesis
CDI is the leading cause of gastroenteritisrelated deaths worldwide. we compared the peptide sequences and compositions from using UPLC-MS/MS. An animal model of CDI was established to investigate the potential therapeutic.
Introduction
Clostridioides difficile (C. difficile) is a Gram-positive, spore-forming, anaerobic bacterium1. It is considered the leading cause of antibiotic-associated diarrhea, accounting for 15–25% of all cases2. It is the causative agent of pseudomembranous colitis associated with antibiotic therapy. In 1935, Hall and O’Toole first described this Gram-positive anaerobic bacillus and named it “difficilis” because of the difficulty of its early isolation and its very slow growth in culture. C. difficile is the leading cause of antibiotic-associated colitis, a disease with significant morbidity and mortality and a major economic burden for hospitalized patients3. The overuse of antibiotics can lead to disturbances in the gut f microbiot, which can lead to an increased risk of CDI4. CDI leads to a range of intestinal disorders with symptoms ranging from mild diarrhea to severe pseudomembranous colitis (PMC), intestinal perforation, which can ultimately be fatal5.
Since 2000, the number and incidence of C. difficile infections have been increasing globally at a high cost, placing a heavy burden on national healthcare systems6. According to data released by the CDC in the United States, in 2021 alone, the gross incidence of CDl was 110.2 cases per 100,000 people, community-related cases were 55.9 cases per 100,000 people, and medical-related cases were 54.3 cases per 100,000 people, causing a heavy financial burden to society. In 2011 alone, C. difficile infected approximately 500,000 people, resulting in approximately 29,000 deaths and an additional $4.8 billion in healthcare expenditures7. A study of 482 hospitals in 20 European countries showed that the average incidence of CDI in 2012–2013 was 7.0 feces per 10,000, with an increasing trend in the overall incidence of CDI in Europe compared to 2005 (2.5 per 10,000) and 2008 (4.1 per 10,000)8. Before 2010, less data related to C. difficile was reported in Asia. From 2013, more research results showed that the incidence of CDI in Asia also showed a rapidly increasing trend9. In China, a study in 2016 showed that among 3953 patients with diarrhea in East China, 397 were diagnosed with CDI, and the clinical symptoms were mainly mild or moderate. The common prevalent strains in the region were ST2, ST3, ST37, and ST54, and the ST37 strain was correlated with clinically severe cases. The study also showed a significant change in the drug resistance of the strains from 200910. In recent years, there has been a rapid increase in the incidence of CDI with increasing mortality and the emergence of many highly pathogenic C. difficile variants11. Therefore, the search for effective treatments for C. difficile infection is urgent.
E. japonicus is a fish with a high annual production worldwide. It has a high nutritional value. The protein content of E. japonicus is 15–20%, which provides rich and cheap protein for developing peptides. However, most E. japonicus are processed into low-value surimi and fishmeal, which are low in value, resulting in a large amount of waste of E. japonicus resources. In recent years, researchers have investigated the activities of peptides from E. japonicus. Zhao12 identified the bioactive peptide PAYCS (Pro–Ala–Tyr–Cys–Ser) from E. japonicus protein hydrolysate and found that different concentrations of PAYCS (PAYCS-L and PAYCS-H) in a scopolamine-induced AD (Alzheimer’s disease) mouse model showed different concentrations of PAYCS (PAYCS-L and PAYCS-H) improve oxidative stress by significantly inhibiting the expression of oxidative indices such as MDA (malonaldehyde) and SOD (Superoxide Dismutase), and alleviate inflammation by reducing the expression of pro-inflammatory factors TNF-α (tumor necrosis factor) and IL-1β (Interleukin-1 beta). Giannetto13 assessed the potential bioactivity of protein hydrolysates obtained from peptides from E. japonicus (APH) by-products in in vitro and in vivo models and demonstrated that APH was effective in modulating the expression of inflammatory factors. In addition, Abbate14 showed that supplementing the diet with 10% (w/w) E. japonicus protein hydrolysate has anti-obesity effects and improves lipid metabolism, reducing hepatic adiposity and liver disease induced by a high-fat diet. However, there is a gap in the current research on peptides from E. japonicus in improving intestinal inflammation, greatly limiting the application and development of E. japonicus in health foods.
G. max contains 40% protein, 20% fat, 10% water, 5% fiber, and 5% ash, making it one of the most nutrient-dense foods. G. max protein has a high absorption rate and can be used as an intestinal nutrient for patients in recovery and in the elderly with declining digestive function. Moreover, G. max peptides can bi-directionally regulate the intestinal environment, promoting microbial growth, reproduction, and metabolism. Kovacs–Nolan15 has shown that G. max tripeptide attenuates the symptoms of colitis and the gene expression of colonic pro-inflammatory cytokines (TNF-α, IL-6, IL-1, IFN-γ, and IL-17). The study found that G. max peptides effectively increased the growth and organic acid secretion of Lactobacillus rohita LR08 and showed synergistic effects with oligofructose in regulating L. rohita LR0816. In addition, it has been shown that fermented barley and a G. max mixture (BS) prevented epithelial barrier dysfunction and increased the level of tight junction proteins in colonic tissues. The leakage of FITC (Fluorescein Isothiocyanate) dextran was evaluated to analyze intestinal permeability, and it was found that BS significantly reduced the leakage of FITC glucan into the blood, indicating that BS intervention effectively enhanced intestinal barrier function. G. max peptides are currently the most commonly used economic peptides in the market for health products. In this study, we analyzed and compared the differences in peptide composition of E. japonicus peptides and G. max peptides using UPLC-MS/MS) coupled with a proteomics database. We also investigated the therapeutic effects of peptides from E. japonicus on antibiotic-induced enteritis and CDI enteritis using peptides from G. max as a control, which provided theoretical support for the novel idea of treating enteritis with natural peptides. In addition, because peptides are small molecules and are easy to absorb, they can provide nutrition for enteritis patients for a long time and help patients with enteritis recover. This study provides ideas and a basis for developing special medical foods for enteritis.
Materials and methods
Materials
E. japonicus peptides were provided by the China Sea Marine Science and Technology Co., Ltd. E. japonicus was provided as a raw material, and it was de-oiled using a press, washed, and desalted. Neutral protease and alkaline protease were used for complex enzymatic hydrolysis. Then, 100 and 800 Da ultrafiltration membranes were used to collect the 100–800-Da components of E. japonicus proteins. G. max peptide was provided by Shandong Tianjiao Biotechnology Co., Ltd., with soybeans as the raw materials. We used water to extract the protein papain, and alkaline protease was used for complex enzyme digestion. We then performed ultrafiltration using a 1000-Da membrane and collected the components that were less than 1000 Da as the G. max peptides. The protein content of the E. japonicus protein and G. max peptides was above 90%, the fat content was 1–2%, and the salt contents were approximately 2.5 and 3.4%, respectively.
Alkaline protease, papain, and neutral protease were purchased from Novozymes Biotechnology Co., Ltd. (China). Cefoperazone, clindamycin, bovine serum protein (66000 Da), cytochrome C (12384 Da), bovine insulin (5733.49 Da), bacteriocins (1422.69 Da), and glutathione (307.32 Da) were purchased from Sigma-Aldrich (St. Louis, MO, USA). LC/MS-grade acetonitrile was obtained from Merck (Darmstadt, Germany). Formic acid (> 98% purity) was purchased from Riedel de Haen (Sleeze, Germany). Vancomycin was purchased from Dalian Meilun Biotechnology Co., Ltd. The real-time polymerase chain reaction (RT–PCR) All-In-One 5X RT MasterMix kit and BlasTaq™2X PCR MasterMix kit were purchased from Applied Biological Materials Inc. (ABM, Canada). PCR primers were purchased from Shanghai Biological Engineering Co., Ltd. The other reagents were analytically grade from Sinopharm Chemical Reagent Co. (Shanghai, China).
Molecular weight and amino acid analysis of the peptides
High-performance size exclusion chromatography (HPSEC) was used to analyze the molecular weights of the peptides on an Agilent 1260 LC system (Agilent CoUSA) equipped with a TSK gel G2000SWXL (300 × 7.8 mm, TosohBioscience, Japan) with acetonitrile/trifluoroacetic acid/water (300/1/700: v/v/v) at flow rate of 0.5 mL/min at 35 °C with a UV detector at 225 nm. The data were processed using GPC software (Agilent, USA). The columns were calibrated by bovine serum protein (66000 Da), cytochrome C (12384 Da), bovine insulin (5733.49 Da), bacteriocins (1422.69 Da), and glutathione (307.32 Da). The weight-average molecular weight (Mw), the number-average molecular weight (Mn), and the polydispersity (Mw/Mn) were calculated using Agilent ChemStation.
2 g of peptide sample was hydrolyzed with 6 mol/L HCl at 110 °C for 24 h in the presence of nitrogen17. The hydrolysate was vaporized by vacuum rotary evaporation at 50 °C, and the residue was dissolved in 25 mL citric acid buffer solution. An aliquot of 0.05 mL was applied to an automated amino acid analyzer (Hitachi-L8900, Japan).
UPLC-MS/MS analysis of peptides
The sequence and contents of peptides in the products were analyzed via UPLC- Peptide mapping were performed using a ULTIMATE 3000 UPLC system (Thermo Fisher Scientific, USA) coupled to Q-Exactive quadrupole-orbitrap ultra-high-resolution mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). An aliquot of 10 μL of peptides were chromatographed on a AdvanceBio Peptide Map column (150 mm × 2.1 mm, 130 Å, 2.7 μm) with a flow rate of 0.25 mL/min and an oven temperature of 40 °C. The mobile phases consisting of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) were programed as follows: 0–2 min, 5% B; 2–27 min, 5–10% B; 27–27 min, 10–25% B; 37–39 min, 20–80% B; 39–43 min,80% B; 43–50 min, 5% B.
The electrospray interface was set in positive ion mode at a resolution of 70,000 with a capillary temperature of 320 °C, a spray voltage of 3.40 kV, a sheath gas flow rate, and an aux gas flow rate at 40 and 10 arbitrary units respectively to obtain the maximum abundance of the fully charged ions in a full-scan spectrum, m/z ranging from 200 to 1500,
The mass spectrometer was operated in positive ion mode at a resolution of 70,000 (MS1 level) with a spray voltage of 3.40 kV, scanning trap capacity of 1 × 105, number of loop count monitored in each acquisition cycle at 20. For ESI-HCD-MS/MS product-ion scanning with 17,500 resolution, a stepped normalized collisional energy of 20, 35, and 40 eV was used with scanning trap capacity of 1 × 105, number of loop count monitored in each acquisition cycle at 20, signal intensity threshold of 1.6 × 105.
The amino acid sequences of all the peptides were obtained by analyzing the data from UPLC-HRMS using the software Peaks Online 1.7 (Bioinformatics Solutions Inc., Canada). The parameters were set as follows. Thresholds for precursor mass tolerance and fragment tolerance were kept at 10 ppm and 0.02 Da, respectively; enzyme: none; missed sites: three; automatic light control (ALC) ≥ 80%.Two databases G. max (NCBI id:3847) and Clupeiforme (NCBI id:32446) were used with ALC ≥ 50% and false discovery rate (FDR) ≤ 1%.
The experimental molecular weight (MExp), theoretical molecular weight (MTheor), and error (ppm) of the peptides were calculated from the single isotope peaks of each peptide. Accurate quantification of each peptide was obtained by extracted ion mass chromatography (EIC) and the peptide peak area was recorded as ion intensity. All quantitative data were normalized to the total identified peptides peak area (in the format of percentage, %).
The peptide compositions of the two peptide products were subjected to Venny analysis (Venny 2.1, http://bioinfogp.cnb.csic.es/tools/venny/index.html).
CDI enteritis modeling
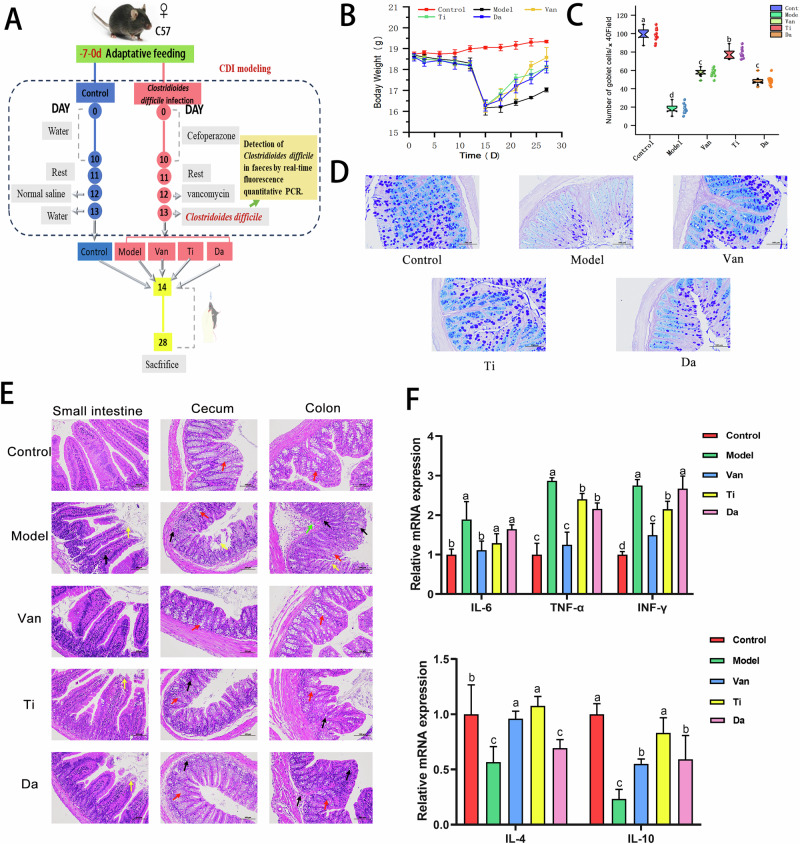
The animal experimental design is shown in Fig. 1A. Forty C57BL/6 mice were acclimatized for 7 days and randomly divided into a normal group (n = 8) and a C. difficile infection group (n = 32). Days 1–10: mice in the normal group were given of ultrapure water daily, and mice in the C. difficile infection group were given with cefoperazone solution at 100 mg/kg of body weight (bw) daily. Day 11: mice in the normal group were injected with saline intraperitoneally, and those in the model group were injected with clindan (50 mg/kg•d). Day 12: the normal group was given with saline at 0.2 mL/day, and the model group was given with C. difficile bacterial solution(1 × 106 CFU/ml) at 0.2 mL. Day 13: the mice rested for 1 day. The weight and state of mice were observed, and the weights of mice in the model group decreased, accompanied by diarrhea, piloerection and huddled. To demonstrate the success of the C. difficile model, the results have been supplemented by the detection of C. difficile in feces by real-time fluorescence quantitative PCR in this study18. The results of the experiment showed that the level of C. difficile in the feces of mice in the model group was 9.12 ± 0.47 (log10CFU/g), indicating that C. difficile had successfully colonized the mice and the CDI model was successfully established.
Fig. 1. Flowchart of animal experiment and Effects of peptides from E. japonicus and G. max on clinical symptoms and intestinal mucosal barrier in mice of CDI.
Different letters indicate significant difference between each group. (n = 5). A Flowchart of animal experiment. B Effects of peptides on the body weight in mice of CDI. C Counting the number of goblet cells in the colon of different groups of mice. D Effects of peptides on colonic Goblet cell in mice of CDI. (×200). E Effects of peptides on pathological changes of small intestine, cecum, and colon tissues in mice of CDI. (×200). Black arrow: inflammatory cell infiltration;Red arrow: goblet cells;Yellow arrow: mucosal epithelial cell erosion and detachment;Green arrow: submucosal edema. F Effects of peptides on colonic inflammatory factors. Pro-inflammatory factors: IL-6(Interleukin 6), TNF-α(Tumor Necrosis Factor-alpha), INF-γ:(Interferon -γ); Anti-inflammatory factors: IL-4(Interleukin 4), IL-10(Interleukin 10). Different lowercase letters represent significant differences between each other for the same indicator (P < 0.05), same as below.
C. difficile infection group (n = 32) was randomly divided into 4 groups: model group (Model group), vancomycin group (Van group), E. japonicus group (Ti group), and G. max group (Da group), with 8 animals in each group. The treatment methods for each group were as follows. The normal and model groups were given with saline of 200 mg/kg bw for the mice. The Van group was given vancomycin of 100 mg/kg bw. The Ti group was given with E. japonicus peptide of 400 mg/kg bw. In the Da group was given with G. max peptide of 400 mg/kg bw. The above test substances were administered orally once a day for two consecutive weeks.
The status of the mice was monitored daily, starting from the day of cefoperazone feeding (100 mg/kg of body weight daily). The mice were observed daily for signs of disease (fecal color, fur color, and mental status), and the daily body mass (g) was recorded. After 14 days of continuous exposure to the formula, the mice were euthanized by Cervical dislocation. The length of the colon was measured, and the contents of the cecum were removed and placed in a sterile tube, which was placed in liquid nitrogen and then transferred to –80 °C for storage for further examination of the intestinal microbiota structure. The small intestine, cecum, and colon tissues were cut into 1 cm each and placed in a 4% formaldehyde solution for soaking for intestinal histopathological analysis. Another 1 cm each of small intestine, cecum, and colon tissue was cut, wrapped in aluminum foil, and then immediately placed in liquid nitrogen for intestinal tissue inflammatory factor detection.
This animal experiment was ethically reviewed by the Specialised Committee on Scientific Ethics of Ocean University of China before it was conducted, and the experiment was carried out in strict accordance with the relevant norms and standards.
Histological analysis of intestinal pathology
In each group, 1 cm2 of small intestine, cecum, and colon tissue samples were taken, fixed with 4% paraformaldehyde for 24 h, embedded, cut into 6-μm sections, and hematoxylin–eosin (H&E) stained19. Tissue sections were coded, randomly grouped, and blinded for scoring using a light microscope.
Colonic goblet cell AB–PA (Periodic acid Schiff and Alcian blue) staining
Paraffin sections of colon tissue were dewaxed in water, stained with a periodate alcohol solution, nucleated with hematoxylin, differentiated by hydrochloric acid alcohol, dehydrated, and sealed with neutral resin. Goblet cells showed red after AB–PAS staining, microscope examination, imaging, and counting the goblet cells.
TUNEL apoptosis fluorescence sections of colon tissue
TUNEL (Terminal Deoxynucleotidyl Transferase mediated dUTP Nick-End Labeling) is a commonly used method for detecting cell apoptosis. We deparaffinized, rehydrated, incubated with histone K, and 3% H2O2 treated colon sections to detect apoptosis in mouse colon epithelial cells. Sections were incubated with terminal deoxyribonucleotidyl transferase (TDT) and 4′, 6-diamidino-2-phenylindole (DAPI) dye solution at 37 °C. Sections were scanned with a laser sc G. ma anning confocal microscope. Cell nuclei were labeled blue with DAPI under UV light, and apoptotic cells were labeled red with CY3. Apoptotic cells were analyzed semi-quantitatively by detecting the relative fluorescence intensity. Relative fluorescence intensity = sum of fluorescence intensities of each area. The relative fluorescence intensity was normalized by the relative fluorescence intensity of the control group.
Colon tissue immunofluorescence analysis
The level of proliferative cell marker PCNA in the colon was detected by the immunofluorescence method to reflect the proliferation of colon epithelial cells. The colon tissue sections were dewaxed, rehydrated, immersed in EDTA buffer (pH = 8.0), and heated in a microwave oven for antigen retrieval. The sections were then blocked with bovine serum albumin and incubated with the primary and secondary antibodies of PCNA and 4′, 6-diamino-2-phenylindole (DAPI) dye solution. The nuclei are labeled blue under UV light with DAPI, and the PCNA is labeled red with CY3. The PCNA expression was semi-quantitatively analyzed by measuring the relative fluorescence intensity of the sections upon scanning with a laser scanning confocal microscope (FV1200, Olympus, Japan). Relative fluorescence intensity = total fluorescence intensity of the region/area of the region. The relative fluorescence intensity of the control group was normalized.
Fluorescent RT–qPCR analysis
The total RNA of mouse colon tissue was extracted according to the instructions of the total RNA extraction kit, and 1 μg of RNA was reverse transcribed into cDNA for each sample. The RT-qPCR system (20 μL) was implemented as follows: 4 μL of cDNA, upstream and downstream primers of 0.6 μL each, 4.8 μL of DEPC water, and 10 μL of Mix. The qPCR system (20 μL) was implemented as follows: 4 μL of cDNA, upstream and downstream primers of 0.6 μL each, 4.8 μL of DEPC (diethyl pyrocarbonate) water, and 10 μL of Mix. The reaction conditions were as follows: pre-denaturation at 95 °C for 10 min; denaturation at 95 °C for 15 s; and annealing/extension at 60 °C for 60 s with 40 cycles in total. The data were analyzed via the 2−ΔΔCT method. GAPDH was used as the internal reference gene correction. The primer sequences were synthesized by Shanghai Sangong Bioengineering Company, and the upstream and downstream primer sequences are shown in Table 1.
Table 1.
Sequences of the primers used in the quantitative RT-qPCR
| Gene Name | Upstream primer sequences (5′-3′) | Downstream primer sequences (5′-3′) |
|---|---|---|
| TNF-α | GCGACGTGGAACTGGCAGAAG | GCCACAAGCAGGAATGAGAAGAGG |
| INF-γ | TCATGGCTGTTTCTGGCTGTTACTG | GACGCTTATGTTGTTGCTGATGGC |
| IL-6 | CTTCTTGGGACTGATGCTGGTGAC | CTCTCTGAAGGACTCTGGCTTTGTC |
| IL-10 | TGGAGCAGGTGAAGAGTGAT | ATTCATGGCCTTGTAGACACC |
| IL-4 | TGGATGTGCCAAACGTCCTC | AGCACCTTGGAAGCCCTACA |
| PCNA | GAGAGCTTGGCAATGGGAACA | ACGTTAGGTGAACAGGCTCATT |
| Ly-6G | CCTTCTCCCAGGATGGACAC | AATTGTAGCACTCCAGCCCC |
| Caspase-2 | ATGGTGATGGTCCTCCCTGTCTTC | CTGTGCGGTCTGGTCATGTAGC |
| Caspase-3 | AGCTTGGAACGGTACGCTAA | CCACTGACTTGCTCCCATGT |
| Caspase-9 | ACAGAGAAGGGGCTTGGG | GCAATAGCATAAAGACAACTCTGG |
| Caspase10 | GTCAAGTTTGCCTACCCCCA | TTCGCAGAAACAGCATTGGC |
| MCM7 | ACCTACCAGCCAATCCAGTCTCC | GAGTAAGCCCTGTGCCATCTGTTG |
| Ki-67 | ACACAGAGCCTTAGCAATAGCAACG | GCCAGTAACACGAGAGTCTTCATCC |
| CyclinD1 | TCTCCTGCTACCGCACAAC | TGGAGGGGGTCCTTGTTTAG |
| ZO-1 | GAAGGCGGATGGTGCTACAAGTG | AGGCTCAGAGGACCGTGTAATGG |
| Occludin | AGTCCACCTCCTTACAGACCTGATG | GCCTCCATAGCCACCTCCGTAG |
| GAPDH | GGTTGTCTCCTGCGACTTCA | TGGTCCAGGGTTTCTTACTCC |
DNA extraction and 16s species diversity analysis of mouse cecum microbiota
The gut microbial structure was examined using a previous method20,21. Genomic DNA from cecum contents was isolated using the PowerFecal™ DNA isolation kit. Then, 2 ng of purified cecum contents of DNA was taken for sequencing and classification. The 16 s rRNA V3-4 region was amplified for PCR using primers 338F/806R. The primer barcodes used were forward: 5′-ACTCCTACGGGGAGGCAGCA-3′ and reverse: 5′-TCGGACTACHVGCACTACHVGGGTWTCTAT-3′18. Paired-end sequencing was performed on an Illumina MiSeq platform provided by Personalbio Technology Co. Sequencing libraries were prepared with Illumina’s TruSeq Nano DNA LT Library Prep Kit.
Analysis of biological information
DNA from the same original DNA was read and merged using FLASH (v1.2.7). Classification units were clustered using Uparse software (v7.0) with 97% similarity, and the classification of each 16S rRNA gene sequence was analyzed with a 70% confidence threshold using the RDP classifier (v2.0). Goods coverage, alpha diversity, and beta diversity were analyzed using Mothur v.1.30.1. Community richness was evaluated using Chao1 and observed species calculations. Shannon and Simpson indices were used to evaluate community diversity, and heat maps based on the OUT relative abundance were generated using Rpackage 2.15. Linear discriminant analysis (LDA) and LDA effect size (LEfSe) analyses were performed using LEfSe software (http://huttenhower.sph.harvard.edu/galaxvUparse/).
Data analysis
All experimental data are expressed as means ± standard deviation. One-way ANOVA was performed using SPSS 25.0 software, and the least significant difference method was used to compare different groups, with P < 0.05 indicating that the difference between them was statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Composition analysis of the peptides
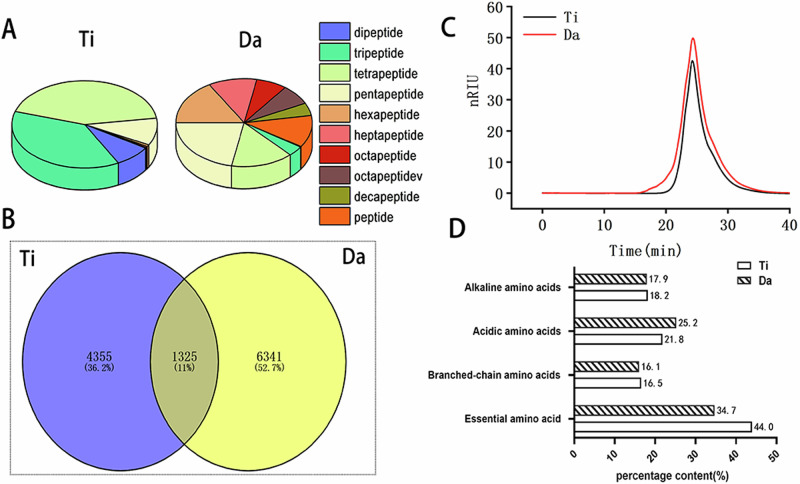
We analyzed the peptide compositions of the two products by UPLC-HRMS. Then, we calculated the peptide sequences and the relative contents of each peptide using Peaks Online 1.7 software. A comparison of the peptide compositions of those tested is shown in Table 2 and Fig. 2A, B. The results showed that the total number of peptides in E. japonicus were 5680. The total number of peptides in G. max were 7666. The total number of peptides in E. japonicus G. max products were 1325, and in E. japonicus the total peptides were dominated by tri and pentapeptides. In addition, the proportion of dipeptides to pentapeptides in E. japonicus was higher (65.5%) than that of the G. max (50.68%). The highest proportion of peptide in G. max was heptapeptide (32.53%), much higher than that of the E. japonicus (3.66%). Tables S1 and S2 show the sequence and MS results of the two peptides, respectively.
Table 2.
Peptide composition analysis of peptides from E. japonicus and G. max
| peptide fragment | Common peptide/chain | E. japonicus peptide composition | G. max peptide composition percentage | ||||
|---|---|---|---|---|---|---|---|
| Specialized peptide/chain | Total ionic strength | Relative amount (%) | Specialized peptide/chain | Total ionic strength | Relative amount (%) | ||
| Amino acids | – | – | 4.34 × 107 | 0.52 | – | 1.82 × 108 | 0.71 |
| Dipeptide | 107 | 33 | 1.64 × 109 | 19.64 | 17 | 3.39 × 109 | 13.19 |
| Tripeptide | 493 | 285 | 2.09 × 109 | 25.04 | 234 | 4.65 × 109 | 18.08 |
| Tetrapeptide | 564 | 986 | 2.24 × 109 | 26.85 | 938 | 5.03 × 109 | 19.56 |
| Pentapeptide | 133 | 1094 | 9.32 × 108 | 11.17 | 1400 | 2.68 × 109 | 10.44 |
| Hexapeptide | 9 | 641 | 3.44 × 108 | 4.12 | 1046 | 1.41 × 109 | 5.49 |
| Heptapeptide | 6 | 444 | 2.92 × 107 | 3.66 | 730 | 8.36 × 109 | 32.53 |
| Octapeptide | 2 | 250 | 2.06 × 107 | 2.58 | 465 | 2.12 × 109 | 8.24 |
| Octapeptidev | 1 | 233 | 2.13 × 107 | 2.67 | 472 | 2.20 × 109 | 8.56 |
| Decapeptide | 0 | 145 | 1.14 × 107 | 1.43 | 291 | 1.63 × 109 | 6.34 |
| Peptide greater than 10 | 0 | 244 | 1.85 × 107 | 2.32 | 748 | 1.22 × 109 | 4.74 |
| Total | 1325 | 4355 | – | – | 6341 | – | – |
Fig. 2. Comparison of the peptide composition of peptides from E. japonicus and G. max.
A Comparison of differences in peptide composition between E. japonicus and G. max peptides. B Wayne analysis of the peptide composition of peptides from E. japonicus and G. max. C Molecular weight analysis of E. japonicus and G. max peptides. D Analysis of the amino acid composition of E. japonicus and G. max.
In addition, the molecular weight and amino acid composition of the two peptides were determined. The molecular weight results of the E. japonicus and G. max are shown in Fig. 2C, D and Table S3. The results showed that the average masses of the E. japonicus and G. max were 264.15 and 413.124 Da, respectively. The results of the amino acid composition (Table 3) showed that the proportions of eight essential amino acids (Lys, Trp, Phe, Met, Thr, Ile, Leu, and Val) in the two protein products (EAA/NEAA) were 44% and 34.7%, respectively. Both peptide composition of different protein products were close to the ideal amino acid pattern defined by FAO/WHO (EAA/TAA: 40%, EAA/NEAA: 60%)22.
Table 3.
Analysis of amino acid composition of peptides from E. japonicus and G.max
| Amino acid name | E. japonicus peptide composition percentage (%) | G. max peptide composition percentage (%) |
|---|---|---|
| Asp | 4.85 | 9.40 |
| Thr | 4.18 | 5.19 |
| Ser | 2.38 | 5.40 |
| Glu | 16.95 | 15.87 |
| Gly | 5.98 | 5.38 |
| Ala | 12 | 3.39 |
| Val | 6.05 | 4.55 |
| Met | 3.42 | 2.04 |
| Ile | 4.29 | 3.43 |
| Tyr | 0.6 | 5.26 |
| Phe | 3.45 | 6.35 |
| Lys | 9.82 | 5.32 |
| Leu | 6.18 | 8.08 |
| His | 5.31 | 5.44 |
| Arg | 3.04 | 7.17 |
| Pro | 2.93 | 3.49 |
| Trp | 6.56 | 2.46 |
| Cys | 1.92 | 1.78 |
The above results showed that the molecular weight, peptide composition, and proportions of the two peptide products differed considerably, supporting the subsequent activity experiments.
Repair of the intestinal barrier in CDI by the two peptides
Peptides repair the intestinal tissue structure
As shown in Fig. 1A, days 1 to 12 of the experiment, mice in other groups except the normal group were given cefoperazone and clindamycin. On the 13th day of the experiment, mice were orally administered 0.2 ml of C. difficile at a concentration of 1 × 106 CFU/mL. The qPCR fluorescence quantification of C. difficile in the feces of mice collected on the first day of infection showed a logarithmic value of C. difficile colonization of 9.12 ± 0.47 (log10CFU/g), indicating that C. difficile had successfully colonized the feces.
Then therapeutic intervention of different peptide products was carried out. The Ti and Da groups were orally administered 400 mg/kg bw per day for two consecutive weeks of E. japonicus peptide and G. max peptide solutions, respectively. At the end of the trial at 2 weeks post treatment, diarrhea gradually stopped, stools became soft, and weight gradually recovered in the treated group compared with the model group.
Figure 1E shows the H&E staining analysis of pathological tissues of small intestine, cecum, and colon of mice in each group. Black arrows indicate inflammatory cell infiltration, red arrows indicate goblet cells, yellow arrows indicate mucosal epithelial cell erosion and detachment, and green arrows indicate submucosal edema. A large number of villous epithelial cells in the mucosal layer of the small intestine in the CDI model group were eroded and detached, and the lamina propria was exposed compared with that of the normal group; the villi became smaller in diameter and inflammatory cell infiltration was seen. In the cecum and colon, some mucosal epithelial cells were eroded and detached, the crypt structure was disturbed, the number of goblet cells was significantly reduced, and a high amount of inflammatory cell influx was seen in the mucosal layer. In contrast, the degree of damage in the Da group was significantly improved compared with the model group, the number of goblet cells was significantly increased, and the degree of inflammatory cell infiltration was reduced. In the Ti group, the degree of damage was further reduced, the number of goblet cells was more, the inflammatory infiltration was further reduced, and the degree of small intestinal epithelial cell necrosis was further reduced.
The number of goblet cells is an important indicator of intestinal barrier function23. After AB–PAS staining of the colon tissue, the goblet cells appeared blue, and basophilic mucins appeared purple. As shown in Fig. 1DC.difficile caused pathological damage to the colon, resulting in reduced goblet cell. The number of goblet cells increased in both the Ti and Da groups. To understand whether the peptides affect the number of colon goblet cells in enteritis mice, we selected 10 random fields to count the goblet cells. As shown in Fig. 1E, compared with the model group, the number of goblet cells in both peptide groups significantly increased, and the number of goblet cells in the Ti group was significantly higher than that in the Da group (P < 0.05).
We analyzed the mRNA relative expression levels of pro-inflammatory and anti-inflammatory cytokines in the colon of mice via RT-qPCR. As shown in Fig. 1F, the mRNA relative expression levels of TNF-α and INF-γ in the colons of the mice in the Ti group were significantly decreased compared with those from the model group (P < 0.05). The relative expression levels of IL-4 and IL-10 mRNA were significantly increased (P < 0.05). In the Da group, the mRNA expression level of TNF-α was significantly decreased, and the mRNA expression level of IL-10 was significantly increased (P < 0.05). These results indicated that both peptides could somewhat inhibit the abnormal increase in the expression of pro-inflammatory factors caused by CDI and promote the expression of anti-inflammatory factors to regulate inflammation. Notably, the effect of the E. japonicus peptide was significantly better(P < 0.05) than that of the G. max peptides.
Peptides regulate the proliferation and apoptosis of colon epithelial cells
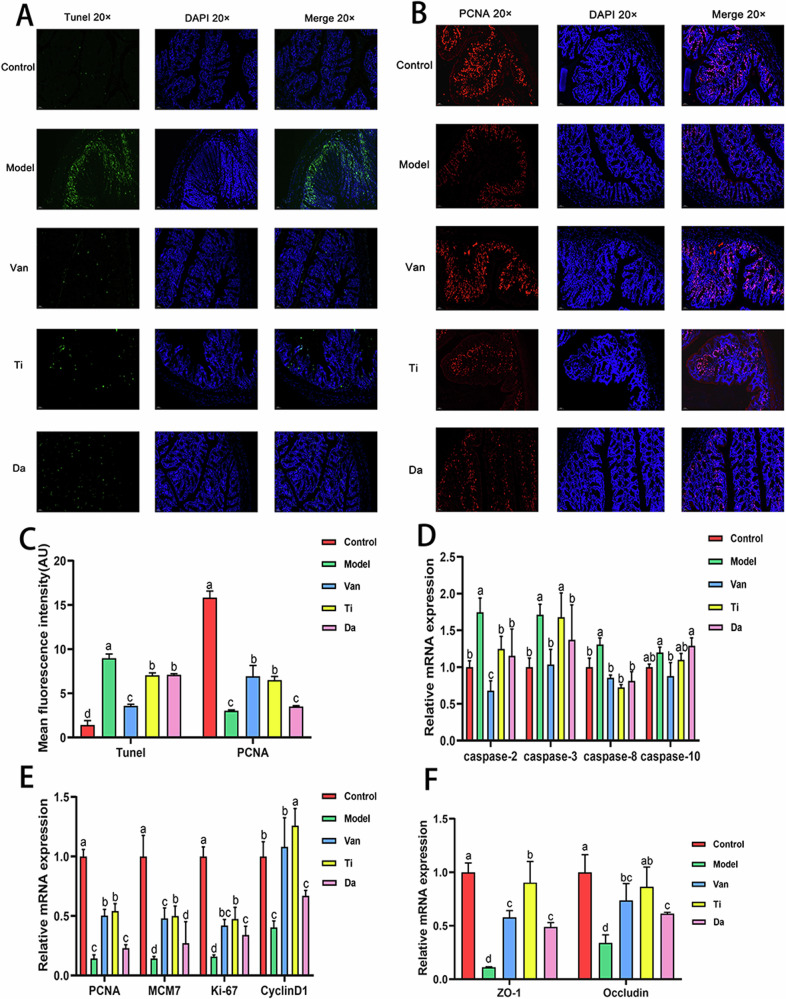
Studies have shown that CDI and the production of TcdA and TcdB can cause apoptosis and death of colon epithelial cells22. Therefore, in this study, the effects of two peptides on apoptosis and proliferation of colonic epithelial cells after CDI were investigated by TUNEL and PCNA fluorescence staining, respectively. As shown in Fig. 3, compared with the model group, the TUNEL fluorescence intensity of the Ti and Da groups decreased by 21.39% and 20.70%, respectively, with no significant difference between the two groups. The fluorescence intensity of the Ti group upon PCNA was significantly increased by 113.53%.
Fig. 3. Effects of peptides from E. japonicus and G. max on apoptosis and proliferation of colonic epithelial cells in mice of CDI.
Different letters indicate significant difference between each group (n = 5). A Colon immunofluorescence staining shows the presence and distribution of apoptotic cells and nuclei, magnification: (B) Existence and distribution of proliferating cells and nuclei, magnification. C Semi-quantitative analysis of fluorescence intensity of TUNEL and PCNA staining. D Effects of peptides on relative expression levels of apoptosis-associated genes in mouse colon epithelial cells. E Effects of peptides on relative expression levels of proliferation-associated genes in mouse colon epithelial cells. F Effects of peptides on the tight junctions of colonic epithelial cells.
Compared with that of the model group (P < 0.05). In comparison, the fluorescence intensity of the Da group only increased by 15.15% compared with the model group (P > 0.05). These results indicated that both products could inhibit the apoptosis of colon epithelial cells, and E. japonicus peptide could effectively promote the proliferation of colon epithelial cells.
In addition, RT-qPCR was used to analyze the relative expression levels of mRNA of caspase-2, caspase-3, caspase-8, and caspase-10, and cell proliferation markers PCNA, MCM7, Ki-67, and CyclinD1 involved in apoptosis (Fig. 3). Regarding apoptosis factors, the mRNA relative expression levels of caspase-2 and caspase-8 of mice in the Ti group significantly decreased (P < 0.05). By contrast, the mRNA relative expression levels of caspase-2, caspase-3, and caspase-8 of mice in the Da group significantly decreased (P < 0.05). There was no significant difference between the two products. For the cell proliferation markers, the mRNA relative expression levels of PCNA, MCM7, Ki-67, and CyclinD1 in the Ti group significantly increased compared with the model group. By contrast, the mRNA relative expression levels of PCNA, MCM7, and CyclinD1 in the Da group were not significantly different compared with those of the model group. The results showed that both groups could inhibit the proliferation of epithelial cells, and E. japonicus peptide could enhance the proliferation of epithelial cells, thus promoting the repair of colon epithelial tissue in mice. This was in good agreement with the previous immunofluorescence staining results.
Peptides promote the tight connection of intestinal epithelial cells
ZO-1 can recognize and transmit various signals, playing an important bridging role in tight junctions7. Occludin is connected to ZO-1 through the C-terminal end, constituting intercellular tight junctions and regulating the cellular paracellular permeability24. The mRNA expressions of tight junction proteins ZO-1 and Occludin were quantitatively analyzed via Reverse Transcription. As shown in Fig. 3F, compared with the model group, mRNA relative expression levels of tight junction proteins ZO-1 and Occludin in the colonic epithelium of the Ti and Da groups significantly increased (P < 0.05). In addition, the expression level of the tight junction protein in the Ti group was significantly higher than that in the vancomycin group (P < 0.05), and the expression level of tight junction protein ZO-1 in the Ti group reached the level of the normal group. This suggests that both E. japonicus and G. max peptides contribute to the close connection of colon epithelial cells, and the repair effect of E. japonicus peptides is significantly higher than that of G. max peptides.
E. japonicus and G. max peptides regulate the intestinal microbiota of CDI
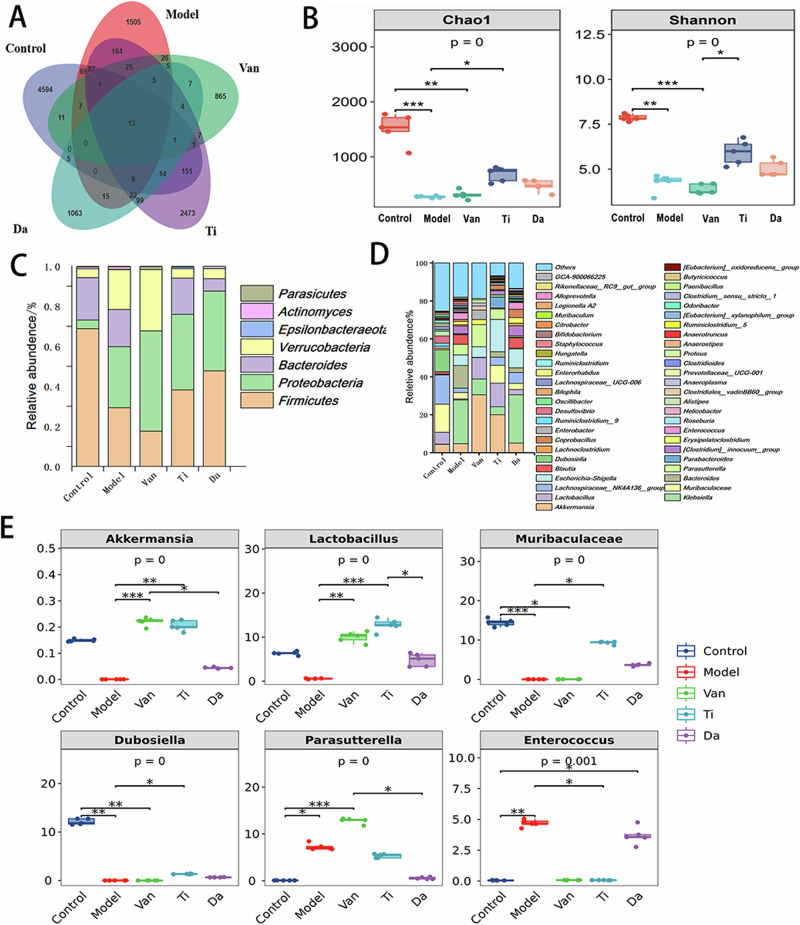
We used 16SrRNA gene sequencing to explore the effects of E. japonicus and G. max peptides on the intestinal microbiota of cecal contents in mice with CDI. After initial screening, quality control, denoising, and correction, a dataset of 2,277,306 high-quality reads was collected for subsequent analysis. As shown in Fig. 4A, the OTUs of the cecum decreased considerably from 4595 to 1505. In addition, Fig. 4B shows that the Chao1 and Shannon indices decreased considerably, indicating that C. difficile infection reduced the number and diversity of bacteria in the cecum. Compared with the model group, the OTUs of the mice in the Ti group increased to 2473, and Chao1 and Shannon indices increased considerably, while the OTUs, Chao1, and Shannon indices of the mice in the Da group did not change considerably. Thus, oral administration of E. japonicus peptides effectively restore the number and diversity of intestinal microbiota of CDI, and its effect is better than that of the G. max peptides.
Fig. 4. Effects of peptides from E. japonicus and G. max on the intestinal microbiota of mice of CDI.
A Effects of peptides on OUTs (Output String) (n = 5). B Effect of peptides on the diversity of bacterial microbiota in the contents of the cecum of mice of CDI. Chao1 richness: colony richness index; Shannon Index: microbiota diversity index (n = 5). C Effects of peptides on the structure of the cecal microbiota in mice of CDI at the phylum level (n = 5). D Effects of peptides on the gut microbiota structure of mice of CDI at the genus level (n = 5). E Effect of peptides on the abundance of beneficial and harmful bacteria. Beneficial bacteria: Akkermansia、Lactobacillus、Muribaculaceae; harmful bacteria: Dubosiella、Parasutterella、Enterococcus (n = 5).
Figure 4C shows the effect of peptides on the structure of the mouse cecum microbiota at the portal level. Five mice from each group were used to calculate the mean relative abundance of intestinal bacteria. After CDI, there was an increase in the proportion of Proteobacteria (30.5%) and a significant decrease in the proportion of Firmicutes (29.29%) in the cecum contents of the mice. Oral administration of E. japonicus and G. max peptides increased the proportion of Firmicutes. It decreased the proportion of Proteobacteria, which resulted in a shift in the microbiota structure of the mice toward the trend of the normal group.
Figure 4D shows a comparison of the effects of E. japonicus and G. max peptides on the structure of intestinal microbiota in the mice analyzed at the genus level. A comparison of the 50 genera with the highest relative abundance revealed that Lactobacillus, Muribacter, Dubosiella, Lactobacillus, and Akkermansia were the dominant bacteria in the cecum of normal mice. Klebsiella, Parasutterella, Blautia, [Clostridioides]_innocuum_group, and Enterococcus became dominant in the cecum of mice after antibiotic treatment and CDI. By contrast, after oral administration of E. japonicus peptides, the intestinal microbial structure of the cecum of intestinally injured mice underwent significant changes. As shown in Fig. 4E, compared with the model group, the contents of beneficial bacteria Akkermansia, Lactobacillus, Muribaculaceae in the cecum of mice in the Ti group were significantly higher, and the proportions of Parasutterella and Enterococcus as harmful bacteria significantly decreased. Akkermansia prevents weight loss, reduces histological damage in the colon, attenuates inflammation, and improves the intestinal barrier in mice, resulting in a protective effect against CDI as a probiotic25. The G. max peptides only significantly increased the proportion of beneficial bacteria Muribaculaceae and decreased the proportion of Parasutterella, and its regulatory effect on beneficial and harmful bacteria was inferior to that of E. japonicus peptides.
Discussion
In recent years, with the emergence of increasing peptide products on the market, they have become a hot research topic. However, the relevant national standards only stipulate the molecular weight and amino acid composition of the peptides present in the market. Currently, LC–MS technology, which combines the powerful separation function of LC with the high sensitivity and selectivity of MS, has gradually become mainstream in peptidomics research. It provides a foundation for researching biological activity and quality control of peptide products. For many years, peptide MS analysis has been characterized by low sensitivity and difficulty in resolving large-volume MS results. The PEAKS online cloud computing platform based on artificial intelligence algorithms has effectively solved these problems26. Studies have shown that the PEAKS online platform resolves 5–30% more peptide sequences than other algorithmic platforms currently leading internationally, and the number of peptides can be re-mined by deep learning prediction with 1.0–1.4 times the number of peptides found previously protein.
E. japonicus and G. max peptides belong to animal and plant proteins, respectively. At present, all peptide products are mixed peptides, and the difference in peptide composition and activity of different protein sources is not clear, so the difference analysis of mixed peptide products was carried out in this study. we first analyzed the contents and sequences of all the peptides in E. japonicus and G. max peptides prepared by enzyme digestion via UHPLC–MS/MS combined with the PEAKS online cloud computing platform. The comparison revealed that the peptide compositions of the two products differed considerably, although the molecular weights were similar. A total of 5680 peptides were found in the E. japonicus sample, and 7666 peptides were found in the G. max sample. The contents of dipeptide to pentapeptide (50.69%) were lower than those of the E. japonicus peptide (65.05%). In comparison, the contents of peptides above 11 amino acid lengths (9.76%) were higher than those of the E. japonicus peptide (4.30%), indicating that the molecular weight of G. max peptides was higher. The number of identical peptides in E. japonicus was 1325, while there were 4355 unique peptides in E. japonicus and 6341 unique peptides in G. max. This indicates that the peptide sequences and composition ratios of the E. japonicus and G. max peptides are also very different, which provides a structural basis for the study to reveal the biological activities of the E. japonicus and G. max peptides. It also provides a reliable quality analysis method for producing peptides with high activity.
C. difficile infection (CDI) establishes in a host, following dysbiosis of the intestinal microbiota, which often occurs following the heavy use of antibiotics to treat unrelated infections, leading to the proliferation of C. difficile within the colon 5. C. difficile’s toxins TcdA and TcdB, can destroy intestinal cells, change the cytoskeleton, and release inflammatory factors, which results in the clinical symptoms associated with C. difficile infection27. In recent years, with the emergence of highly virulent strains of C. difficile, the morbidity and mortality of C. difficile infections have increased significantly around the world, and the severity of the disease has also risen markedly, resulting in more patients entering the ICU, resection of the colon, and even death, among other serious consequences28. Therefore, the search for an effective treatment for C. difficile infection is urgent.
Changes in the intestinal microbiota caused by conventional antibiotic therapies can create an intestinal environment suitable for C. difficile overgrowth, exacerbating the severity of CDI and even causing CDI recurrence7. Probiotics, as a novel therapeutic modality, are effective in altering intestinal microbiota, antimicrobial activity, intestinal barrier protection, and immunomodulation. Currently, probiotic supplementation has attracted more and more clinical attention in the prevention and treatment of CDI patients24. In addition, food-borne fucoidan can also effectively regulate intestinal microbiota and help CDI recovery29. As small molecules, peptides are easily absorbed. They can provide long-term nutrition to patients with enteritis and aid recovery. Thus, there have been numerous studies on the therapeutic role of peptides in enteritis in recent years. Studies have shown that a variety of food-derived peptides play an active role in the treatment of enteritis, including regulating intestinal microbiota, reducing intestinal inflammation, and repairing the intestinal mucosal barrier7,24,27–29. Therefore, the present study aimed to investigate whether food-borne peptides, including E. japonicus and G. max peptides, can effectively repair the barrier damage, reduce intestinal inflammation, and regulate the disordered intestinal microbiota in CDI, in order to fill the gaps in this study, and to provide a theoretical basis for the subsequent research on C. difficile enterocolitis-related speciality foods.
However, in recent years, E. japonicus peptides, a resourceful marine fish with a high nutritional value, have primarily been processed into low-value products, such as feed and surimi. There is a lack of studies on the therapeutic effects of E. japonicus peptides on enteritis. In addition, because E. japonicus and G. max peptides differ dramatically in terms of specific peptide composition, it is necessary to explore what differences exist in terms of the biological activity of these two different sources of peptides.
Therefore, in our study, a CDI model was used to investigate the therapeutic effects of E. japonicus and G. max peptides on CDI. Both peptides were able to repair the intestinal mucosal barrier, reduce intestinal inflammation, and promote the proliferation of intestinal epithelial cells and the production of tight junction proteins. However, E. japonicus peptides were better than G. max peptides, especially in promoting the proliferation of colonic epithelium and the maintaining of goblet cells, which may be because E. japonicus peptides contain a higher proportion of small molecule peptides in their composition. We further explored the effects of the two peptides on the structure of the intestinal microbiota of CDI mice, which showed large differences. The E. japonicus peptides significantly increased the richness and diversity of the microbiota. They significantly increased the abundance of beneficial intestinal bacteria, such as Akkermansia, Lactobacillus, and Muribaculaceae. They lowered the number of harmful bacteria, shifting the intestinal microbiota structure to normal conditions. This study provides a theoretical basis for developing healthy foods with E. japonicus peptides as the main functional ingredient and special foods for enteritis.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript. Funding This work was supported by the National Key Research and Development Program of China (grant No. 2021YFC2301000), the National Sci-Tech key project (2018ZX10733402), and the National Key Research and Development Program of China (grant No. 2021YFC2301000).
Author contributions
Ying Li, Yuan Wu, and Xue Zhao designed the study and performed the data analyses. Ying Li and Yuan Wu performed methodology, data analysis, visualization, and wrote the draft preparation. Ying Li, Yan zhe Li, Lu lu Bai, Ya jun Jiang, Zhan Wang, and Te long Xu confirmed the data and revised the manuscript. Yuan Wu and Xue Zhao supervised the study and contributed to review and editing the manuscript. All authors approved the submitted version.
Peer review
Peer review information
Communications Biology thanks Romain Villéger and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Sabina Leanti La Rosa and Joao Valente. A peer review file is available.
Data availability
The datasets presented in this study can be found in supplementary Data 1 and supplementary Data 2.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuan Wu, Email: wuyuan@icdc.cn.
Xue Zhao, Email: zhaoxue@ouc.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06850-x.
References
- 1.Lawson, P. A., Citron, D. M., Tyrrell, K. L. & Finegold, S. M. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe40, 95–99 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Simpson, H. L. et al. Soluble non-starch polysaccharides from plantain (Musa x paradisiaca L.) diminish epithelial impact of Clostridioides difficile. Front. Pharmacol.12, 766293 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly, C. P., Pothoulakis, C. & LaMont, J. T. Clostridium difficile Colitis. N. Engl. J. Med330, 257–262 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Heuler, J., Chandra, H. & Sun, X. Mucosal Vaccination Strategies against Clostridioides difficile Infection. Vaccines11, 887 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor, J. R., Johnson, S. & Gerding, D. N. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology136, 1913–1924 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Hudson, S. L. et al. Probiotic use as prophylaxis for Clostridium difficile-associated diarrhea in a community hospital. Am. J. Infect. Control47, 1028–1029 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Barker, A. et al. Probiotics for Clostridium difficile infection in adults (PICO): study protocol for a double-blind, randomized controlled trial. Contemp. Clin. Trials44, 26–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, K. A. et al. Underdiagnosis of Clostridium difficile across Europe: the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID). Lancet Infect. Dis.14, 1208–1219 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Khun, P. A. & Riley, T. V. Epidemiology of Clostridium (Clostridioides) difficile Infection in Southeast Asia. Am. J. Trop. Med. Hyg.107, 517–526 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin, D. et al. Molecular epidemiology of clostridium difficile infection in hospitalized patients in Eastern China. J. Clin. Microbiol.55, 801–810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman, J. et al. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev.23, 529–549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao, T. et al. PAYCS alleviates scopolamine-induced memory deficits in mice by reducing oxidative and inflammatory stress and modulation of gut microbiota-fecal metabolites-brain neurotransmitter axis. J. Agric. Food Chem.70, 2864–2875 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Giannetto, A. et al. Protein hydrolysates from anchovy (Engraulis encrasicolus) waste: in vitro and in vivo biological activities. Mar. Drugs18, 86 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbate, J. M. et al. Administration of protein hydrolysates from anchovy (Engraulis Encrasicolus) waste for twelve weeks decreases metabolic dysfunction-associated fatty liver disease severity in ApoE–/–Mice. Animals10, 2303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovacs-Nolan, J. et al. The PepT1-transportable soy tripeptide VPY reduces intestinal inflammation. Biochim. Biophys. Acta1820, 1753–1763 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Zhu, Y., Chen, G., Diao, J. & Wang, C. Recent advances in exploring and exploiting soybean functional peptides-a review. Front. Nutr.10, 1185047 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M., Liu W. & Li G. Isolation and characterisation of collagens from the skin of largefin longbarbel catfish (mystus Macropterus). Food Chem.115, 826–831 (2009).
- 18.Chu, Q. et al. Detection of Clostridium difficile with TaqMan-based quantitative RT-PCR[J]. Dis. Surveill.33, 417–422 (2018). [Google Scholar]
- 19.Reeves, A. E. et al. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes2, 145–158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi, H. et al. Dietary fucoidan of Acaudina molpadioides alters gut microbiota and mitigates intestinal mucosal injury induced by cyclophosphamide. Food Funct.8, 3383–3393 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Wang, L. et al. Fucoidan isolated from Ascophyllum nodosum alleviates gut microbiota dysbiosis and colonic inflammation in antibiotic-treated mice. Food Funct.11, 5595–5606 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Li, Z. et al. Amino acid profiles and nutritional evaluation of fresh sweet–waxy corn from three different regions of China. Nutrients14, 3887 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birchenough, G. M. H., Johansson, M. E., Gustafsson, J. K., Bergström, J. H. & Hansson, G. C. New developments in goblet cell mucus secretion and function. Mucosal Immunol.8, 712–719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oksi, A., Anttila, V.-J. & Mattila, E. Treatment of Clostridioides (Clostridium) difficile infection. Ann. Med.52, 12–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasiri, G. et al. The inhibitory effects of live and UV-killed Akkermansia muciniphila and its derivatives on cytotoxicity and inflammatory response induced by Clostridioides difficile RT001 in vitro. Int. Microbiol.27, 393–409 (2023). [DOI] [PubMed]
- 26.Xin, L. et al. A streamlined platform for analyzing tera-scale DDA and DIA mass spectrometry data enables highly sensitive immunopeptidomics. Nat. Commun.13, 3108 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandrasekaran, R., Kenworthy, A. K. & Lacy, D. B. Clostridium difficile toxin a undergoes clathrin-independent, PACSIN2-dependent endocytosis. PLoS Pathog.12, e1006070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debast, S. B., Bauer, M. P. & Kuijper, E. J. European Society of Clinical Microbiology and Infectious Diseases. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin. Microbiol. Infect.20, 1–26 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Luo, J. et al. A comparative study of the effects of different fucoidans on cefoperazone-induced gut microbiota disturbance and intestinal inflammation. Food Funct.12, 9087–9097 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The datasets presented in this study can be found in supplementary Data 1 and supplementary Data 2.