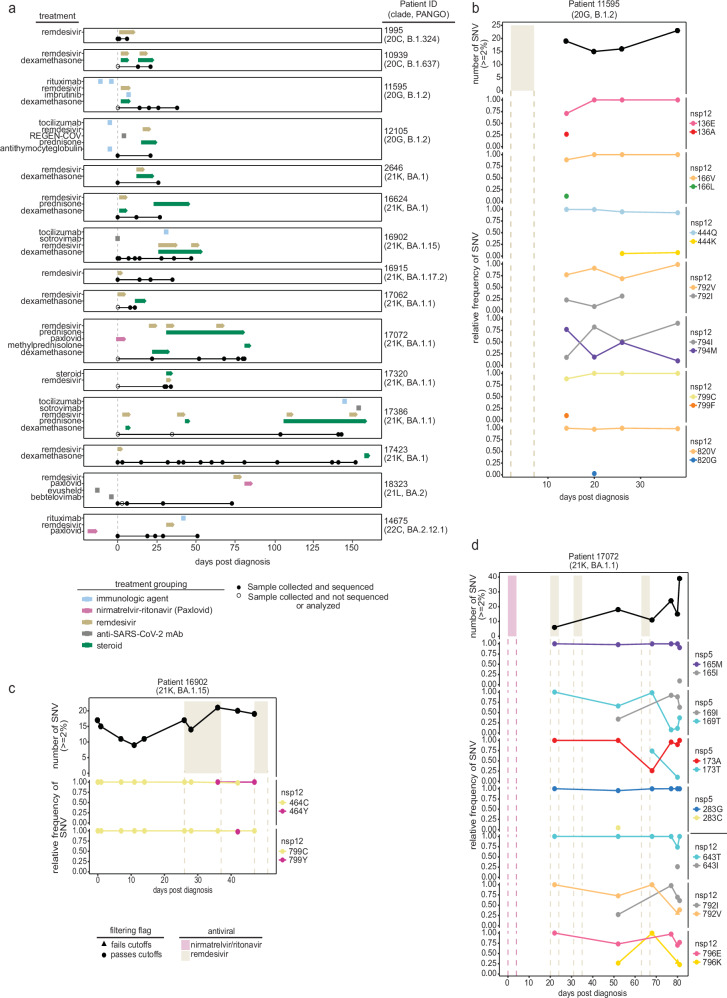
Fig. 1. Emergence of SARS-CoV-2 variants in immunocompromised patients.
a The treatment course and sample collection from 15 patients with persistent SARS-CoV-2 infections. Clade and PANGO lineage designations are included with Patient IDs. All time points along the x axis are referenced from the date of each patient’s initial positive COVID-19 test result, with day 0 marking the date of the first positive test at the New York Presbyterian (NYP) Hospital (dashed gray line). Treatment courses are specified along the y axis for each patient and colored based on the treatment category. Nasopharyngeal swab samples successfully sequenced and used for analyses are represented as black points along the solid black line for each patient. The total number of unique single-nucleotide variants (SNVs) found ≥2% across the genome (top) over the course of the infection in patients b 11595, c 16902, and d 17072 compared to their treatment courses with nirmatrelvir-ritonavir (pink) or remdesivir (tan). SNVs are determined by comparing each sample to their respective clade consensus sequences (“Methods”). The relative frequency of non-synonymous SARS-CoV-2 SNVs in nsp5 and nsp12 coding regions during infection are shown below. Variant data are grouped by the coding region, amino acid position, and amino acid residue, with the color representing the amino acid and the shape indicating whether the variant was found above (passes, circle) or below (fails, triangle) our detection cutoffs (“Methods”). All time points along the x axis are referenced from the date of each patient’s initial positive COVID-19 test result, with day 0 marking the date of the first positive test at the NYP hospital.