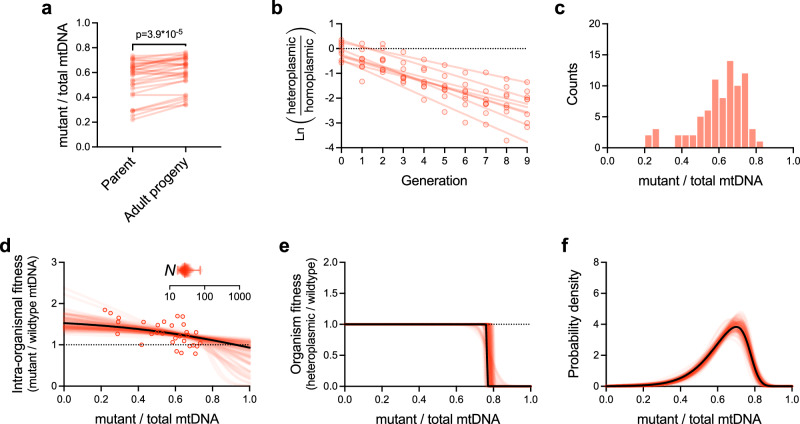
Fig. 3. Integration of benchtop experimentation with theoretical modeling reveals the stable maintenance of a selfish mitochondrial genome via a combination of multilevel selection and genetic drift.
a–c Empirical data (provided in Source Data file). a Pairwise comparisons of mutant frequency (uaDf5) between parent and progeny for the purpose of measuring intra-organismal selection on a selfish mitochondrial genome (n = 30 parent-progeny lineages). Two-tailed Wilcoxon matched-pairs test. b Inter-organismal selection against uaDf5, measured by directly competing heteroplasmic carriers of the selfish genome against homoplasmic (wildtype) animals on the same food plate, in a mixed population (n = 8 competed populations). Data shown as the log of the heteroplasmic over homoplasmic fraction of each replicate competed population (see Methods); regression lines are fit to the data from each replicate population by least squares. c histogram of mutant frequencies of age-synchronized adults (n = 81) sampled from a heteroplasmic stock population (same data as in Fig. 1b). Data in a–c are taken from our prior study26. d–f Maximum-likelihood model of the intra-organismal (d) and inter-organismal (e) fitness effects, each as a function of mutant frequency, and the most evolutionarily stable mutant frequency distribution (f). Each plot in d–f shows the maximum-likelihood estimates corresponding to the empirical data in a–c (solid black lines in d–f) plus 100 simulated data sets for parametric bootstrapping (red lines) to visualize the confidence estimates for each plot. Model parameters specifying intra-organismal selection, inter-organismal selection, and intra-organismal genetic drift (d, inset) were collectively estimated using a joint maximum-likelihood approach that combines all data sets in (a–c). Error bars in genetic drift plot (d, inset) indicate mean, minimum, and maximum bootstrap values. See Supplementary Data 1 for empirical and bootstrap model parameters.