Figure 2 |. Livers of patients with localized PaC exhibit molecular alterations with prognostic significance.
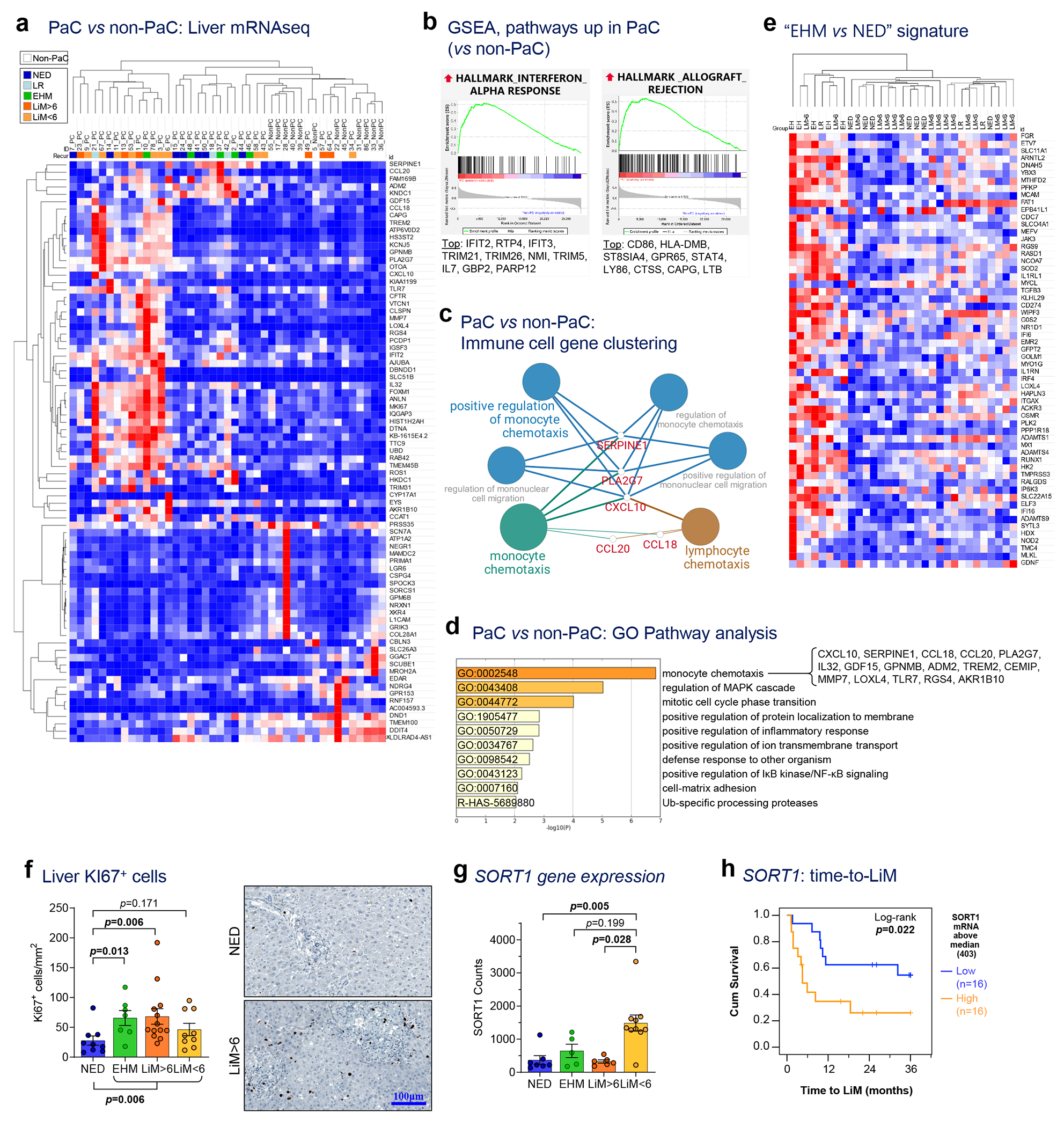
a, mRNA sequencing of liver tissue collected intraoperatively identified 79 genes differentially expressed between pancreatic cancer (PaC; n=31) and non-PaC patients (n=12; Wald test performed using the DESeq2 package with adjustment for multiple comparisons; genes shown were altered >2-fold, with adjusted p<0.1). PaC patients were classified into five mutually-exclusive recurrence groups: No evidence of disease (NED); isolated local recurrence (LR); extrahepatic metastasis (EHM); early liver metastasis (within 6 months, LiM<6); and late LiM (beyond 6 months, LiM>6). b, Enriched gene sets related to immune response in PaC livers by GSEA using the Hallmarks of Cancer reference gene set (MSigDB, H; pathways considered significant if p<0.05, FDR<0.25). The top 10 genes driving each gene set are listed, in descending order. c, Immune cell gene clustering and visualization of genes significantly upregulated in PaC livers by Cytoscape ClueGO. d, Pathway gene expression analysis of significantly upregulated genes by Metascape (cutoff p<0.1, after adjustment for multiple comparisons). e, Unsupervised clustering using the genes differentially expressed between EHM and NED patients and f, Confirmatory Ki-67 immunostaining showing upregulation in recurrence groups (n=38; Mean±SEM; Kruskal-Wallis ANOVA p=0.023, pair-wise testing with correction for multiple comparisons shown if p<0.25). g,h, SORT1 expression in recurrence groups, showing g, upregulation in LiM<6 (n=29; Mean±SEM; Kruskal-Wallis ANOVA p=0.002, pair-wise testing with correction for multiple comparisons shown if p<0.25); h, association with time to LiM (TTLiM) (n=32; log-rank test; p=0.022).
