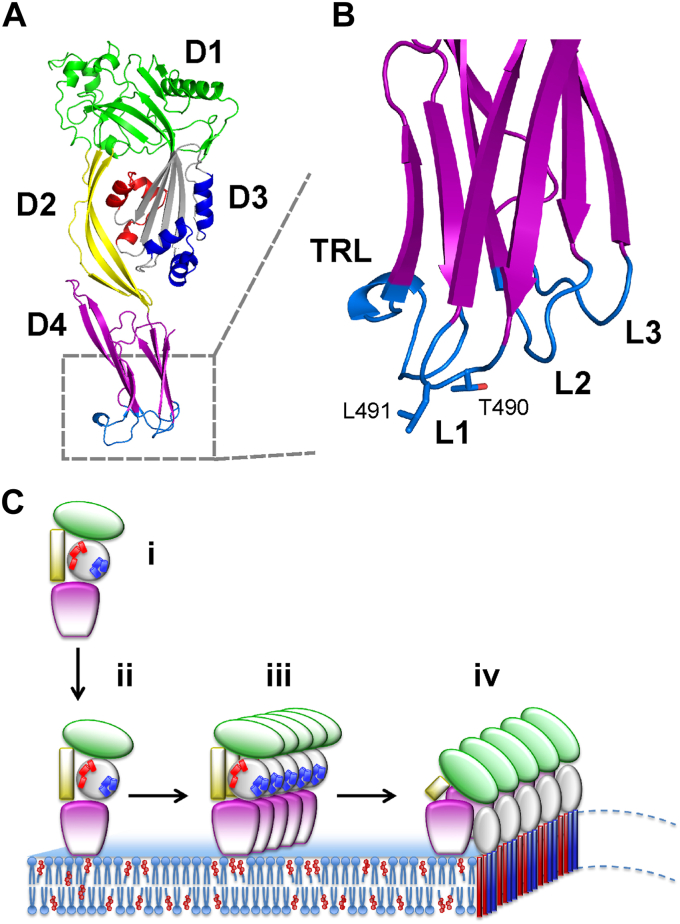
Figure 1.
3D structure of PFO and mechanism of pore formation.A, a 3D model of the structure of PFO (PDB ID: 1PFO (29)). Different domains are labeled and colored. Two clusters of α-helices in D3 that participate in pore formation are labeled in red and blue. B, enlargement of the lower part of D4. Loops 1 to 3 (L1–L3) and the tryptophan-rich loop (TRL) comprising membrane-binding surface are labeled. The conserved residues Thr490-Leu491 implicated in cholesterol recognition (CRM) are shown in sticks. The view is rotated 90° clockwise in comparison to panel A. C, pore formation by PFO proceeds through several steps: binding of the monomer (i) to the lipid membrane (ii), oligomerization of the monomers in the plane of the membrane (iii) and insertion of the beta strands of D3 into the lipid membrane to form the final pore (iv). Labeling of different parts of a protein is as in (A). Images of the 3D models were created in Pymol (61).