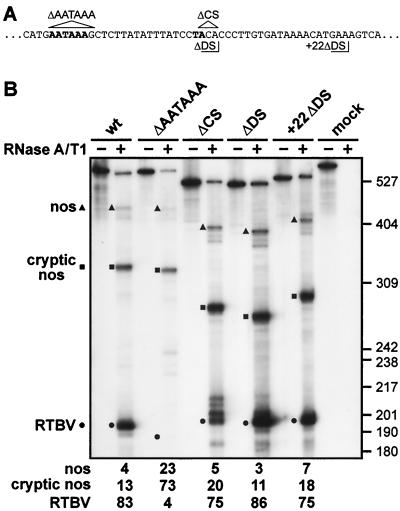
FIG. 2.
AATAAA is required for 3′-end processing; the cleavage site and specific downstream sequences are not. (A) Sequences surrounding the AATAAA and cleavage sites (both in bold type): nucleotides deleted in ΔAATAAA and ΔCS are indicated by triangles; the end points of RTBV sequences in ΔDS and +22ΔDS are delimited by right-angled lines. (B) RNase protection assay to examine processing events at the RTBV poly(A) site in a wild-type construct (RTPA-2), and constructs carrying deletions of either AATAAA (ΔAATAAA), the cleavage site (ΔCS), or sequences directly downstream of the cleavage site (ΔDS) or from 22 nt downstream of the cleavage site (+22ΔDS) as shown in panel A. Protected fragments corresponding to transcripts processed at the RTBV, cryptic nos, and nos sites are indicated, with the positions of size markers (pBR322/HpaII) shown on the right. Processing efficiencies (in percent) are given underneath. The gel shown is from analysis of RNA from N. plumbaginifolia protoplasts.