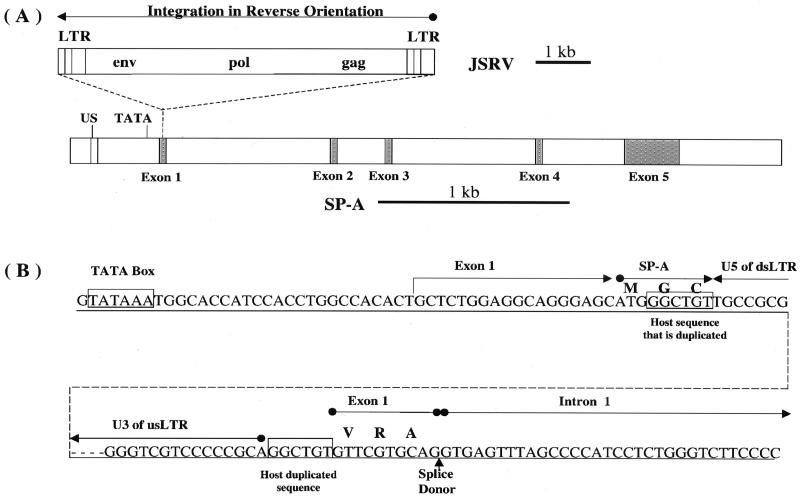
FIG. 6.
SP-A is the site of JSRVJS7 proviral integration. (A) JSRVJS7-provirus is integrated in a reverse orientation to the 5′ untranslated region of SP-A exon 1. Both JSRVJS7 and SP-A sequence fragments are shown to scale. Ovine SP-A exons are shaded and introns are white. US is a 25-nucleotide upstream “enhancer” region which shares high homology to sequences in human and baboon. (B) The boundaries of the integration site are shown in detail. Exon 1 is predicted to begin approximately 25 nucleotides downstream of the TATA box. Integration of JSRVJS7 interrupts the proposed six-amino-acid exon 1 SP-A translation product. The dotted line represents the JSRVJS7 provirus. GGCTGT is the retroviral target sequence duplicated by the host upon integration. Amino acids are indicated above some of the codons in the nucleotide sequences as follows: M, Met; G, Gly; C, Cys; V, Val; R, Arg; A, Ala. us, upstream; ds, downstream.