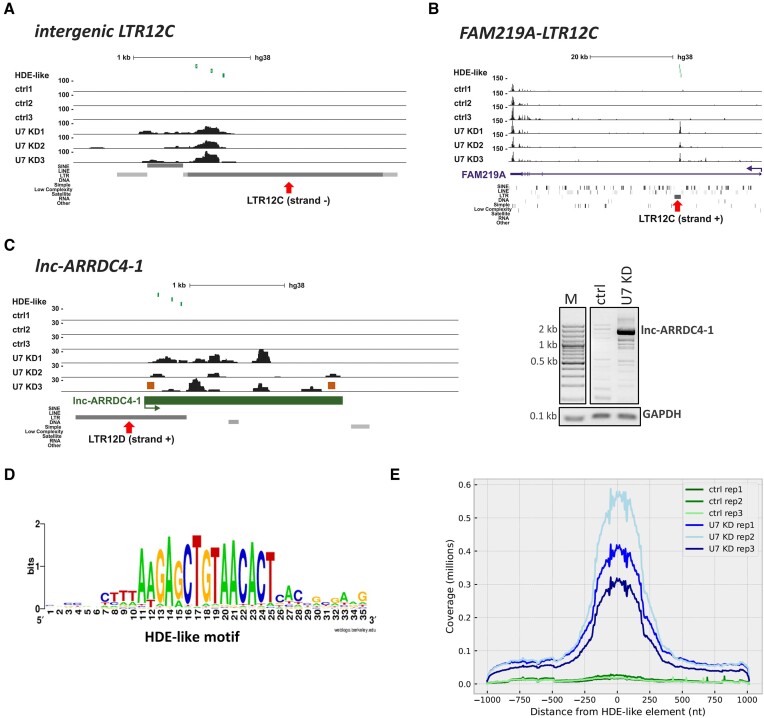
Figure 2.
U7 snRNA regulates the expression of LTR12s and lincRNAs containing HDE-like motifs. (A–C) UCSC Genome Browser visualization of RNA-seq data from HEK293T ctrl and U7 KD cells (three biological replicates are shown) of the intergenic LTR12C element (A), the intragenic LTR12C element in FAM219A (B) and the LTR12-containing lincRNA lnc-ARRDC4-1 (C). Peaks within data tracks illustrate reads mapped to particular genomic regions. The positions of HDE-like motifs are shown in the top track as green vertical lines. The red arrows indicate the position of the LTR12 elements within the gene structures. The expression of full-length lnc-ARRDC4-1 was checked by semiquantitative PCR using primers depicted as orange boxes, and GAPDH was used for normalization (C, on the right). (D) Consensus sequence of the HDE-like motif present within HERVs in the context of the 10 nucleotides flanking the motif on both sides. (E) Graph showing the distribution of reads around HDE-like motifs within LTR12 elements present in the human genome. The Y axis represents the cumulative number of reads assigned to a corresponding position around the HDE-like elements, which are placed at position ‘0′.