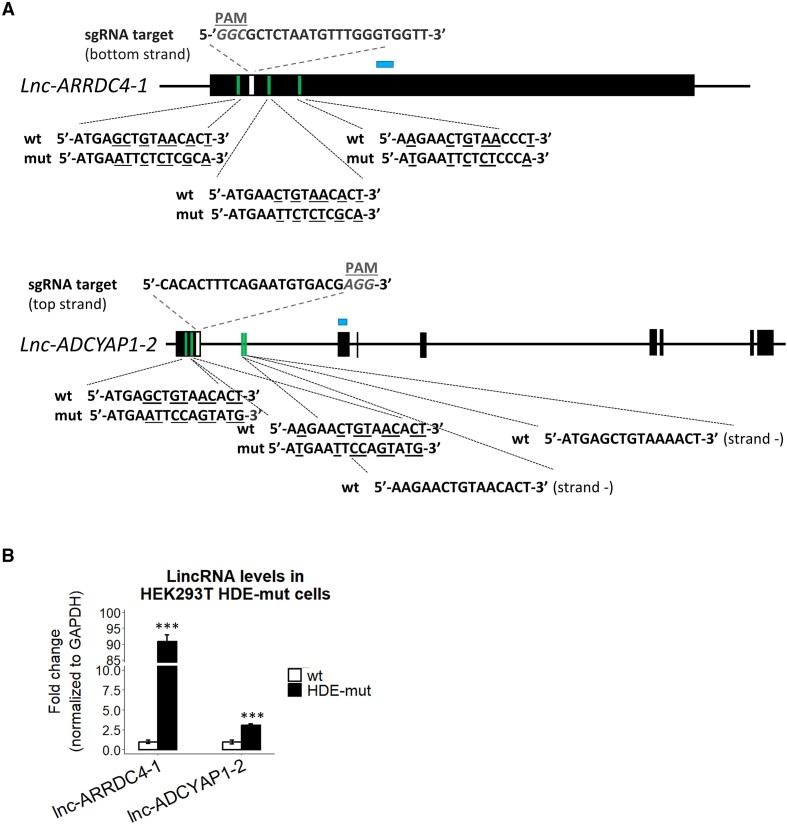
Figure 3.
U7 snRNA controls the expression of LTR12s and lincRNAs via HDE-like motifs. (A) Schematic representation of lnc-ARRDC4-1 and lnc-ADCYAP1-2 (isoform ENST00000670380.1). The black boxes represent exons, and the lines depict introns. The positions of HDE-like motifs and sgRNA target sequences used for the generation of HEK293T lnc-ARRDC4-1 HDE-mut and lnc-ADCYAP1-2 HDE-mut cells using the CRISPR−Cas9 system are indicated in green and white, respectively. Nucleotides mutated in the HDE-like motifs are underlined. HEK293T lnc-ARRDC4-1 HDE-mut cells are heterozygous, with one allele corresponding to the wild-type versions of HDE-like motifs and the other carrying desired mutations within HDE-like motifs. Two HDE-like motifs located within the intron of the lnc-ADCYAP1-2 gene (in antisense orientation) were not edited. PAM, protospacer-adjacent motif. (B) The expression levels of the respective lincRNAs in lnc-ARRDC4-1 HDE-mut and lnc-ADCYAP1-2 HDE-mut cell lines were analyzed by RT−qPCR. The locations of the amplified qPCR products in the expression analysis are shown as blue boxes in (A). GAPDH levels were used for normalization. Data are presented as means ± SDs (n = 3). P values were calculated using two-sided Student's t test, and statistical significance was defined as follows: ***P ≤ 0.001.