Abstract
Emerging data have highlighted a correlation between microbiome composition and cancer immunotherapy outcome. While commensal bacteria and their metabolites are known to modulate the host environment, contradictory effects and a lack of mechanistic understanding impede the translation of microbiome-based therapies into the clinic. In this study, we demonstrate that abundance of the commensal metabolite pentanoate is predictive for survival of chimeric antigen receptor (CAR) T cell patients in two independent cohorts. Its implementation in the CAR T cell manufacturing workflow overcomes solid tumor microenvironments in immunocompetent cancer models by hijacking the epigenetic-metabolic crosstalk, reducing exhaustion and promoting naive-like differentiation.
While synergy of clinically relevant drugs mimicked the phenotype of pentanoate-engineered CAR T cells in vitro, in vivo challenge showed inferior tumor control. Metabolic tracing of 13C-pentanoate revealed citrate generation in the TCA cycle via the acetyl- and succinyl-CoA entry points as a unique feature of the C5 aliphatic chain. Inhibition of the ATP-citrate lyase, which links metabolic output and histone acetylation, led to accumulation of pentanoate-derived citrate from the succinyl-CoA route and decreased functionality of SCFA-engineered CAR T cells. Our data demonstrate that microbial metabolites are incorporated as epigenetic imprints and implementation into CAR T cell production might serve as embodiment of the microbiome-host axis benefits for clinical applications.
Introduction
Although approval of CAR T cell therapies has marked a major breakthrough in the therapy of hematologic neoplasms, many patients with advanced malignancies and non-hematologic cancers in particular do not benefit from these. This is in part due to T cell-intrinsic dysfunctionality or suppressive factors within the tumor microenvironment (TME) [1, 2]. Despite all current efforts, these hostile influences in solid tumors are diminishing the efficacy of CAR T cells in patients, underscoring the necessity for new, interdisciplinary sources of improvement. The host’s immune environment determines the success of immunotherapies which can be influenced by the intestinal microbiome both under homeostatic and pathophysiological conditions [3–5]. Its impact on cancer immunotherapy was first recognized in the context of immune checkpoint inhibitor therapy (ICI) when several studies demonstrated that response and resistance to PD-1 and CTLA-4 blockade depends on the gut microbial composition [5, 6]. Colonization with Akkermansia muciniphila and some Bifidobacterium strains facilitates the improvement of ICI via activation of endogenous T and antigen-presenting cells [6, 7]. These cells also translocate from the intestine to the tumor site, thereby inspiring the design of commensal consortia to modulate T cell response favorably [8]. Recently, the importance of the microbiome in chimeric antigen receptor (CAR) T cell therapy was highlighted by correlation of antibiotic treatment prior to CAR T cell infusion with worse response and progression free survival (PFS) [9, 10]. While microbial metabolites and producer strains influence anti-tumor response, their modes of action often remain obscure. In this regard, short-chain fatty acids (SCFAs; a major group of commensal molecules) have been investigated in inflammation, autoimmunity and cancer [11–14]. Contradictory findings highlight their context- and cell type-dependent effects raising a need for both: a detailed mechanistic understanding and decisive strategies to apply commensal metabolites for clinical use. We have previously demonstrated that pentanoate is a commensal metabolite that augments the anti-cancer efficacy of CD8 T cells [15].
In this study, we identified a strong correlation between abundance of pentanoate and progression-free survival (PFS) of CAR T cell patients, suggesting a potential impact on the apheresis product and CAR T cell fitness. We have developed a protocol which integrates pentanoate-mediated epigenetic-metabolic reprogramming into the CAR T cell manufacturing workflow in order to confer superior in vivo function, metabolic fitness and a naive-like cell state, overcoming and modulating the detrimental TME in immunocompetent solid tumor models. Although the synergy of clinically available modulators mocetinostat and dichloroacetate (DCA) mimicked the metabolite’s HDAC-inhibitory and glucose oxidative effects in vitro, in vivo application of CAR T cells manufactured with these drugs demonstrated inferior tumor control as compared to pentanoate-engineered cells. Metabolic tracing of 13C-labeled pentanoate revealed that the C5 molecule uniquely hijacks the cellular metabolism by feeding into the tricarboxylic acid (TCA) cycle via two different entry points, contributing to citrate translocation into the nucleus and consequently histone acetylation. These findings show that commensal metabolites can shape the T cell’s epigenetic imprint and confer features that cannot be mirrored by clinically available drugs. The implementation of pentanoate into the CAR T cell manufacturing workflow has the potential to optimize adoptive T cell therapy and serves as an example to exploit the microbiome-host interactions for precision design of next generation anti-cancer drugs.
Results
PIM exposure confers adverse progression-free and overall survival
We recently have identified the human microbiome strain Megasphaera massiliensis as a potent producer of the SCFAs pentanoate. Its strong capacity to induce anti-tumor features in T cells prompted us to investigate its role in a clinical real-life context.
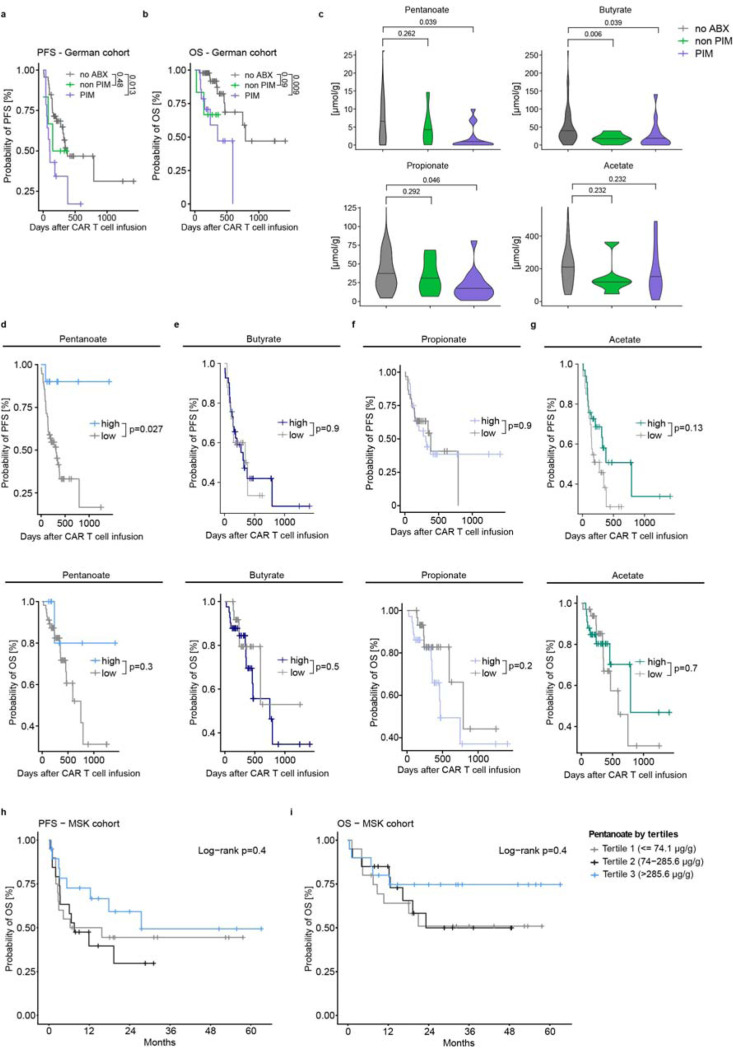
Previous reports highlighted the impact of broad-spectrum antibiotics on anti-CD19 CAR T cell therapy outcomes [9, 10]. Consistently, in our cohort described in Suppl. Table 1, exposure to PIM group antibiotics (piperacillin/tazobactam, imipenem, meropenem) prior to (any target) CAR T cell therapy resulted in worse PFS (HR 2.66, Fig. 1a) and OS (HR 4.11, Fig. 1b) compared to patients not exposed to antibiotics. Conversely, patients receiving non-PIM antibiotics showed similar PFS (HR 1.55, Fig. 1a) and only a trend towards adverse OS (HR 3.95, Fig. 1b) compared to those not treated with antibiotics.
Figure 1: Pentanoate abundance is a predictor for clinical CAR T cell response.
a and b, Probability of progression-free survival (a) and overall survival (b) in a German cohort of patients receiving CAR T cells, depending on different antibiotic-(ABX)-treatment regimen. PIM (piperacillin/tazobactam, imipenem, meropenem); PFS, progression-free survival; OS, overall survival. c, Levels of multiple short-chain fatty acids following antibiotic exposure. d-g, Kaplan-Meier curves showing PFS (upper panel) and OS (lower panel) depending on the abundance of pentanoate (d), butyrate (e), propionate (f) and acetate (g). h,i Kaplan-Meier curves showing PFS (h) and OS (i) depending on the abundance of pentanoate in a US cohort.
Patients with PIM exposure display reduced fecal pentanoate levels
Examination of the association between SCFA levels and antibiotic exposure revealed that pentanoate levels were significantly reduced only in patients with PIM exposure (Fig. 1c). No significant reduction was observed with non-PIM antibiotics. Similar effect could be observed with the odd-chain family member propionate. However, levels of the even-chain fatty acid butyrate, were already decreased with non-PIM antibiotics. Acetate showed no significant reduction with either PIM or non-PIM antibiotics. These correlations suggest that PIM exposure substantially affects pentanoate-producing commensal and thereby PFS.
Pentanoate levels prior to CAR T cell infusion associate with progression-free survival
Next, we evaluated the relationship between fecal SCFA levels and PFS of CAR T cell patients. Patients with high levels of pentanoate exhibited significantly better PFS compared to those with low levels of pentanoate (HR 6.9, estimated 1-year PFS 90% vs. 41.5%; median PFS not reached vs. 310 days, Fig. 1d). These patients also exhibited a trend towards improved overall survival (HR 3.0, estimated 1 year OS 80% vs. 71.7%, median OS not reached vs. 749 days, Fig. 1d). Notably, other SCFAs, such as butyrate, propionate and acetate could not predict PFS and OS to a similar degree (Fig. 1e-g). In another cohort of patients (n=60) with non-Hodgkin lymphoma treated with CD19-directed CAR T cells (Suppl. Table 2), pre-CAR-T infusion fecal pentanoate concentrations were categorized by terciles. The higher terciles had numerically higher 2-year PFS (p=0.4) and 2-year OS (p=0.4, Fig. 1 h-i). Thus, pentanoate abundance might serve as a predictive parameter for CAR T cell response in patients.
Implementation of pentanoate in the CAR T cell manufacturing improves product characteristics.
Based on the results of the correlative analysis, we hypothesized that the superior PFS might be a consequence of pentanoate-mediated immunostimulation improving host environment and CAR T cell function. To investigate the latter and develop an approach to exploit pentanoate’s benefits for clinical application facing hostile, immunosuppressive TME, we designed a protocol featuring a short in vitro expansion phase designed to better retain stemness and fitness of the CAR T cell product in the host after infusion (Fig. 2a).
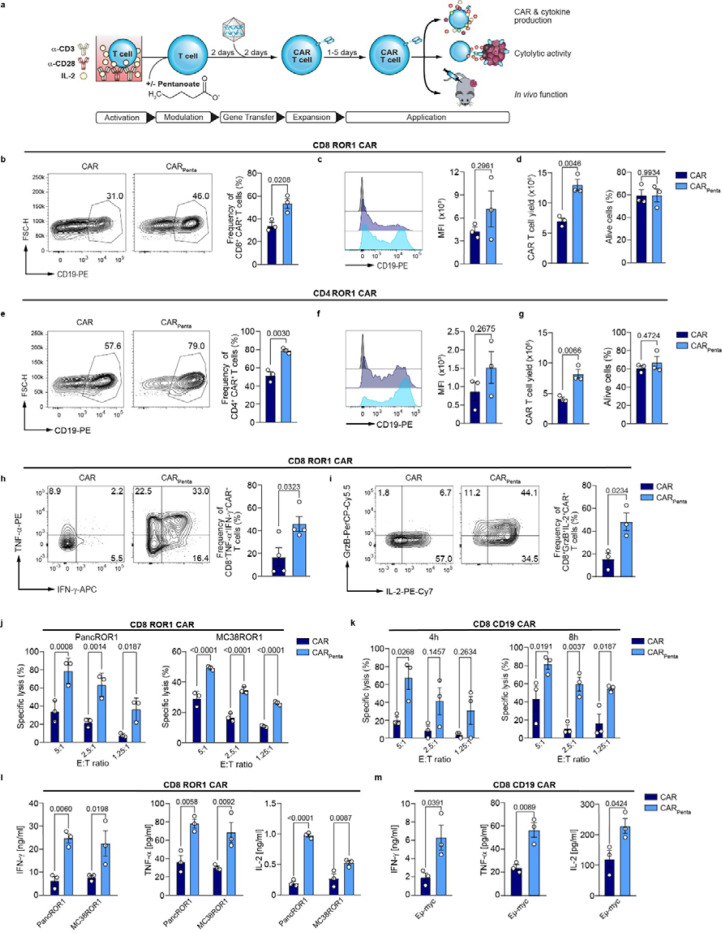
Figure 2: Pentanoate improves engineering and favors CTL-phenotype of murine CAR T cells.
a, Schematic illustration depicting CAR T cell generation with pretreatment and functional analysis. b, Representative contour plots of the transduction marker and bar plots show frequencies of CD8+ ROR1 CAR T cells on day 7. Mean ± SEM from n = 3. c, Median fluorescence intensity (MFI) of CAR+ T cells. Mean ± SEM from n = 3. d, Total CAR+ T cell yield and viability on day 7. Mean ± SEM from n = 3. e, Representative contour plots and bar graphs show transduction efficiency of CD4+ CAR T cells on day 7. Mean ± SEM from n = 3. f, Median fluorescence intensity (MFI) for generated CAR+ T cells. Mean ± SEM from n = 3. g, Yield and viability of CAR T cells on day 7. Mean ± SEM from n = 3. h, Frequency of IFN-γ and TNF-α producing ROR1 CAR T cells on day 4 following antigen-independent restimulation for 5 h. Mean ± SEM from n = 3. i, Flow cytometric analysis of Granzyme B and IL-2 production by CD8+ ROR1 CAR T cells after restimulation on day 4. Mean ± SEM from n = 4. j, Specific cytolytic activity of ROR1 CAR T cells against tumor cells at different E:T ratios after 6 h. Mean ± SEM from n = 3. k, Killing capacity of CD19 CAR T cells measured by specific lysis of CD19-expressing tumor cells at different E:T ratios after 4 and 8 h. Mean ± SEM from n = 3. l and m, Cytokine secretion of IFN-γ, TNF-α and IL-2 after 24 h co-culture of ROR1- (l) or CD19- (m) specific CAR T cells with target cells. Mean ± SEM from n = 3.
(b-i) Data represent pooled data from independent experiments. (j-m) n=3 biological replicates; pooled data from n=3 independent experiments; mean ± SEM was calculated for n=3 independent experiments. Statistical analysis was performed using unpaired two-tailed Student’s t test (b-i) and two-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test (j-m).
Isolated T cells from murine spleens and lymph nodes were activated with α-CD3 and α-CD28 antibodies in presence of pentanoate for 2 days, prior to retroviral gene transfer of the CAR transgene. Between 3 to 7 days post transduction, we characterized phenotypic and functional changes both in vitro and in vivo (Fig. 2a-m). Generation of CD4+ and CD8+ ROR1-specific CAR T cells showed that both transduction efficacy and transduction marker expression were elevated in the pentanoate-engineered CAR T cells (CARPenta) as compared to the untreated ones (CAR), further accompanied by increased cell yield without impairment of viability (Fig. 2b-g). Similar data were obtained when the workflow was applied to the production of CD4+ and CD8+ CD19 CAR T cells (Extended Data Fig. 1a-f). Moreover, both ROR1 and CD19 CARPenta T cells showed strong upregulation of the effector cytokines TNF-α and IFN-γ as well as granzyme B and IL-2 in an antigen-independent manner (Fig. 2h, i and Extended Data Fig. 1g, h). Prior to assessment of cytotoxic activity, CAR T cells were cultured for 4 days without pentanoate to explore the longevity of the modulation. Co-cultures of ROR1-specific CARPenta T cells with the ROR1-expressing murine pancreatic ductal adenocarcinoma cells Panc02 (PancROR1) or colorectal carcinoma cells MC38 (MC38ROR1) as well as of CD19-specific CARPenta T cells with CD19+ E μ-myc cells showed significant improvements in cytotoxic activity and antigen-dependent cytokine release as compared to conventionally engineered CAR T cells (Fig. 2j-m and Extended Data Fig. 1i-l). These results suggest that short-term pentanoate treatment leads to a sustained phenotype in different CAR products.
HDAC class I-inhibition-mediated hyperacetylation enhances CAR T cell function
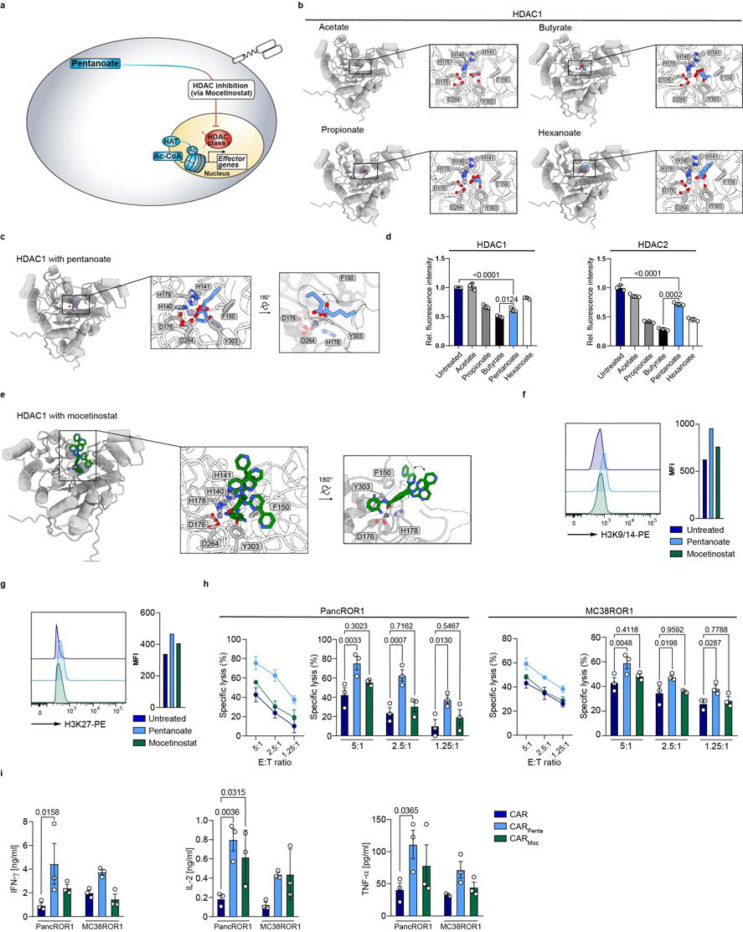
The sustained increase in effector function after removal of the commensal metabolite suggests a lasting effect of pentanoate. This might be attributed to SCFA-mediated inhibition of histone deacetylases (HDACs) and consequently histone hyperacetylation, thereby altering the epigenetically programmed fate of eukaryotic cells (Fig. 3a, b). To predict how HDACs interact with fatty acids, we used AlphaFold 3 to model zinc and palmitate bound structures of classes I and II HDACs (Extended Data Fig. 2a-c). The structural prediction revealed that HDACs of both classes coordinate palmitate in their reactive centers with high confidence up to C5 (pLDDT >90 for class I and 70–90 for class II; Extended Data Fig. 2d). Docking of C2-C6 SCFAs (acetate to hexanoate) into an AlphaFold 3-predicted structure of zinc cofactor-bound HDAC1 suggested that the aliphatic chains might bind in an inward facing orientation up to a chain length of C4 and in an outward facing conformation for C6 SCFAs (Fig. 3b, c and Extended Data Fig. 3a). Notably, docking of pentanoate (C5) showed two conformations, with the best scoring inward facing conformation being preferred (predicted ΔG ≈ −8.6 kcal/mol), relative to the best outward facing orientation (predicted ΔG ≈ −7.6 kcal/mol, Extended Data Fig. 3a). Docking of pentanoate into HDACs of class I and IIa suggestedpreferred binding to class I enzymes (Extended Data Fig. 3b, c). Experimentally, we confirmed the predicted inhibition of HDAC1 and 2 over class IIa by SCFAs (Fig. 3d and Extended Data Fig. 3d). Although the SCFAs analyzed are all structurally similar and elicit HDAC inhibition, their effects are highly context- and cell type-dependent. Differentiation of CD4 T cells towards regulatory T cells (Tregs) has long been linked to the HDAC-inhibitory activity of SCFAs [3, 13, 16]. Interestingly, only propionate and butyrate, but not pentanoate, induced the Treg master regulator Foxp3 (Extended Data Fig. 4). Haradhvala and colleagues have reported that patient relapse is associated with the presence of CAR Tregs, favoring the use of pentanoate over other SCFAs [17]. Of note, treatment of inducible Tregs (iTregs) with pentanoate suppressed Foxp3 and reciprocally induced the CTL/ Th1-related transcription factor T-bet in a concentration-dependent manner (Extended Data Fig. 5a-d). Tregs derived from T-bet-deficient (Tbx21−/−) animals were unable to express the same level of IFN-γ as compared to their WT counterparts. Moreover, engineering of CAR T cells in the presence of different SCFAs highlighted pentanoate as the most potent modulator of anti-tumor activity (Extended Data Fig. 6a, b). These findings suggest that use of pentanoate could repress CAR Treg development and favor CTL/ Th1 polarization.
Figure 3: HDAC class I inhibition-mediated hyperacetylation enhances CAR T cell function.
a, Schematic pathway illustration detailing the HDAC-mediated T cell regulation. b and c, Shown are SCFAs (blue sticks), i.e. acetate, propionate, butyrate, pentanoate and hexanoate docked into AlphaFold 3-predicted models of zinc cofactor-bound (spheres) HDAC1 (gray cartoons). Three highest scoring conformations are shown superimposed for each ligand. d, Influence of bacterial SCFAs on the activity of recombinant class I and class II HDAC enzymes. e, Mocetinostat (green sticks, HDAC class I inhibitor) docked into an AlphaFold 3-predicted model of zinc cofactor-bound (spheres) HDAC1 (gray cartoons). Three highest scoring conformations are shown superimposed. f and g, Histone acetylation status of T cells are probed by the expression of H3K9/14 (f) and H3K27 (g) via flow cytometry after treatment with indicated substances. One representative experiment is shown (n = 3). h, Specific cytolytic activity of CD8+ CAR T cells generated in the presence of indicated substances against ROR1-expressing tumor cell lines at different E:T ratios after 6 h. Mean ± SEM from n = 3. i Cytokine secretion of CD8+ CAR T cells after 24 h co-culture with target antigen expressing cell lines, measured for IFN-γ, TNF-α and IL-2 by ELISA. Mean ± SEM from n = 3.
h and i, biological replicates; pooled data from independent experiments. Statistical analysis was performed using one-way ANOVA (h) or two-way ANOVA with Tukey’s multiple-comparison test (i).
To assess whether clinically relevant HDAC class I inhibitors could mimic pentanoate’s modulatory benefits, we engineered ROR1 CAR T cells in presence of mocetinostat (CARMoc). AlphaFold 3-modeling of zinc-bound HDAC1 and docking of mocetinostat into the reactive center suggests a binding mode similar to that of pentanoate, with a predicted ΔG of approximately −8.1 kcal/mol (Fig. 3e and Extended Data Fig. 3a). Intracellular flow cytometry staining for histone post-translational modifications revealed an increase in acetylation within CD8 and Th1 T cells at H3 Lys9/14 and H3 Lys27, respectively, both prominent marks of open chromatin and transcriptional activity (Fig. 3f, g and Extended Data Fig. 8a). Next, we analyzed the consequences of pentanoate- and mocetinostat-mediated hyperacetylation for the CD4 and CD8 CAR T cell phenotype. Surprisingly, and despite mocetinostat’s strong HDAC-inhibitory activity, CARMoc T cells elicited less potent lysis of target cells and antigen-specific cytokines secretion compared to CARPenta T cells, although both were superior in killing relative to untreated CAR T cells (Fig. 3h, i). Engineering in the presence of HDAC class II inhibitor TMP-195 did not improve CAR T cell features (Extended Data Fig. 8b), in line with our previous findings. These results suggest that epigenetic remodeling affects the CTL phenotype; however, a standalone implementation of a clinical HDACi such as mocetinostat is not sufficient to fully reconstitute CARPenta T cells features.
Pentanoate synergizes epigenetic-metabolic modulation to boost effector function.
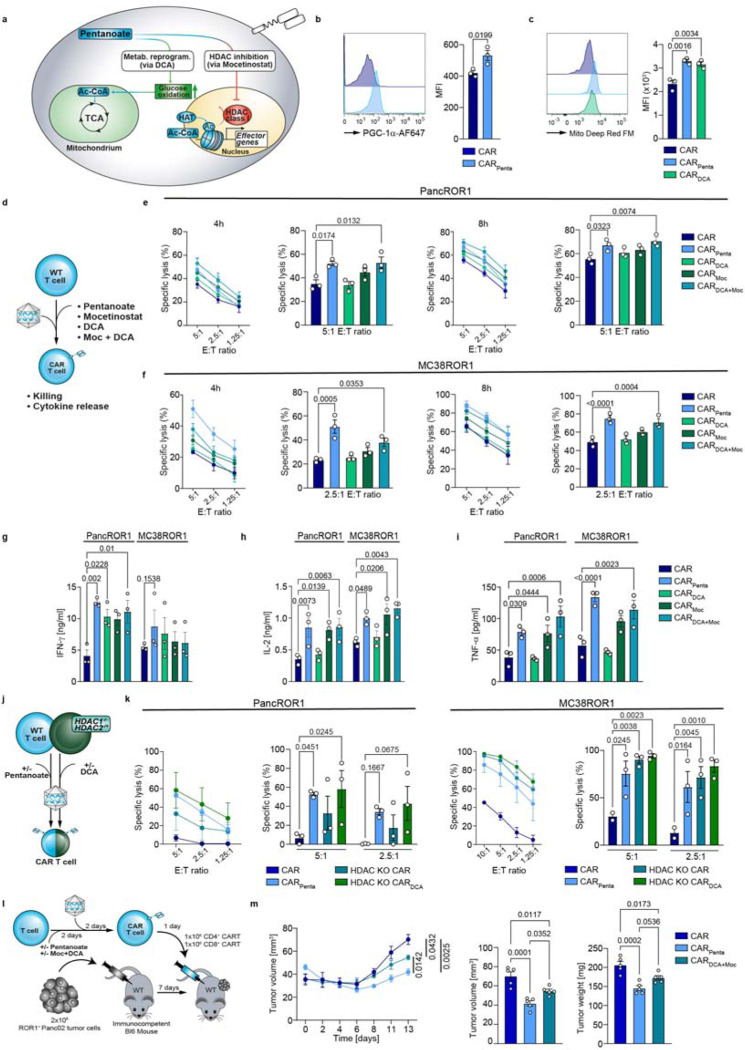
We next investigated whether pentanoate influences cellular metabolism to augment HDACi function (Fig. 4a). CD4 and CD8 CARPenta T cells showed increased phosphorylation of the central metabolic regulator S6, a downstream target of mTOR (Extended Data Fig. 9a, b). By staining metabolic key functions, we observed an increased glucose and fatty acid uptake (Extended Data Fig. 9c, d). Moreover, CD4 and CD8 CARPenta T cells increased the expression of the master regulator of mitochondrial biogenesis PGC-1α and mitochondrial mass, compared to control CAR T cells (Fig. 4b, c and Extended Data Fig. 9e, f). Thus, an mTOR-mediated shift towards glucose oxidation might contribute to the CARPenta T cell phenotypeTo probe this effect in a more isolated manner, we next generated CAR T cells in presence of the clinically used drug dichloroacetate (DCA, CARDCA T cells), according to our initial concept. Of note, DCA is a pharmacological inhibitor of mitochondrial pyruvate dehydrogenase kinase (PDK) and is currently under clinical investigation for cancer therapy [18]. PDK activates pyruvate dehydrogenase (PDH), which itself acts as a gatekeeper enzyme for pyruvate flux into the TCA cycle. Consequently, DCA redirects metabolism from lactate fermentation to glucose oxidation in mitochondria. Similar to pentanoate, engineering of CARDCA T cells increased fatty acid uptake and mitochondrial mass (Fig. 4c and Extended Data Fig. 9d). Thus, we decided to use DCA to mimic the glucose oxidative enhancement during the engineering process.
Figure 4: Pentanoate synergizes epigenetic and metabolic modulation to boost effector function.
a, Schematic pathway representation. b, Flow cytometry analysis and quantification of PGC-1α in CD8+ CAR T cells either untreated or treated with pentanoate. Mean ± SEM from n = 3. c, Flow cytometric analysis of MitoFM in CD8+ CAR T cells treated with indicated substances. Mean ± SEM from n = 3. d, Combinatorial or single treatment workflow for CD8+ T cells. e and f, Cytolytic activity of ROR1 CAR T cells against PancROR1 (e) and MC38ROR1 (f) at different E:T ratios after 4 h and 8 h. Mean ± SEM from n = 3. g-j, Secretion of IFN-y (g), IL-2 (h) and TNF-α (i) after 24 hours by CD8+ CAR T cells upon co-incubation with ROR1-expressing tumor cell lines. Mean ± SEM from n = 3. j, Scheme illustrating the different pretreatments of CAR+ T cells generated from wildtype or HDAC knockout mice. k, Cytolytic activity of CD8+ CAR T cells against target cells with different E:T ratios after 4 h. CAR T cells were generated as shown in j. Mean ± SEM from n = 3. l, Experimental setup of pretreatment with pentanoate or DCA/mocetinostat combination during the CAR+ T cell manufacturing process followed by injection in PancROR1 tumor bearing mice. m, Tumor growth (left) over time as well as tumor volume (middle) and tumor weight on day 14 (right) after tumor inoculation are shown. Mean ± SEM from n = 10 mice/group until day 7, then n = 5 mice/group until day 14.
b, c, e-l and k, independent experiments. e-l, k, biological replicates; pooled data from independent experiments. Statistical analysis was performed using one-way ANOVA (b, c, m) or two-way ANOVA with Tukey’s multiple-comparison test (e-I and k).
Next, we tested whether the simultaneous HDAC inhibition and metabolic reprogramming synergistically improved CARPenta T cell effector function. To do so, we engineered CAR T cells in the absence or presence of pentanoate, DCA, mocetinostat or a combination of the latter, respectively (Fig. 4d). The engineered T cells were subjected to co-cultures using PancROR1 or MC38ROR1 tumor cells to assess cytotoxicity (Fig. 4e, f). Interestingly, while CARDCA T cells did not show a significant improvement in killing, combined administration of DCA and mocetinostat during manufacturing (CARDCA+Moc T cells) resulted in an additive effect which was also reflected in antigen-dependent secretion of TNF-α, IFN-γ+ and IL-2 (Fig. 4g-i). Noteworthy, varying the co-stimulatory domain within CAR constructs alters the cellular metabolism and therefore CAR T cell function [19]. We thus performed a set of experiments using a CD28-based ROR1 CAR. This resulted in similar effects upon treatment with the previously mentioned drugs with regards to cytotoxicity and mitochondrial mass (Extended Data Fig. 10a, b).
To understand a potential contribution of mocetinostat off-target effects, we probed HDAC class I involvement by using mice T cells with homozygous HDAC1- (HDAC1−/−) and heterozygous HDAC2-(HDAC1−/−) deficiency (Fig. 4j). HDAC-deficient CAR T cells showed superior specific lysis compared to the WT CAR control, mimicking the trends observed in CARMoc T cells (Fig. 4k). Engineering in the presence of DCA further boosted lytic activity and cytokine secretion, comparable to levels of CARPenta T cells (Fig. 4k and Extended Data Fig. 11). This demonstrates a synergistic relationship between HDAC inhibition and metabolic enhancement that appears to be responsible for the CARPenta T cell phenotype.
To evaluate whether the in vitro results could be recapitulated in vivo, we applied suboptimal doses of CAR, CARPenta and CARDCA+Moc T cells in an immunocompetent pancreatic tumor model characterized by a cold TME, without lymphodepletion (Fig. 4l). Surprisingly, differences in tumor mass and a lack of peripheral expansion indicated that the clinical drugs were not able to reproduce the efficacy of pentanoate, although improved tumor control was observed in both groups as compared to the control CAR T cells (Fig. 4m and Extended Data Fig. 12a, b). Thus, though mocetinostat and DCA mimic some key features of pentanoate, other mechanisms might be involved that are not affected by these drugs.
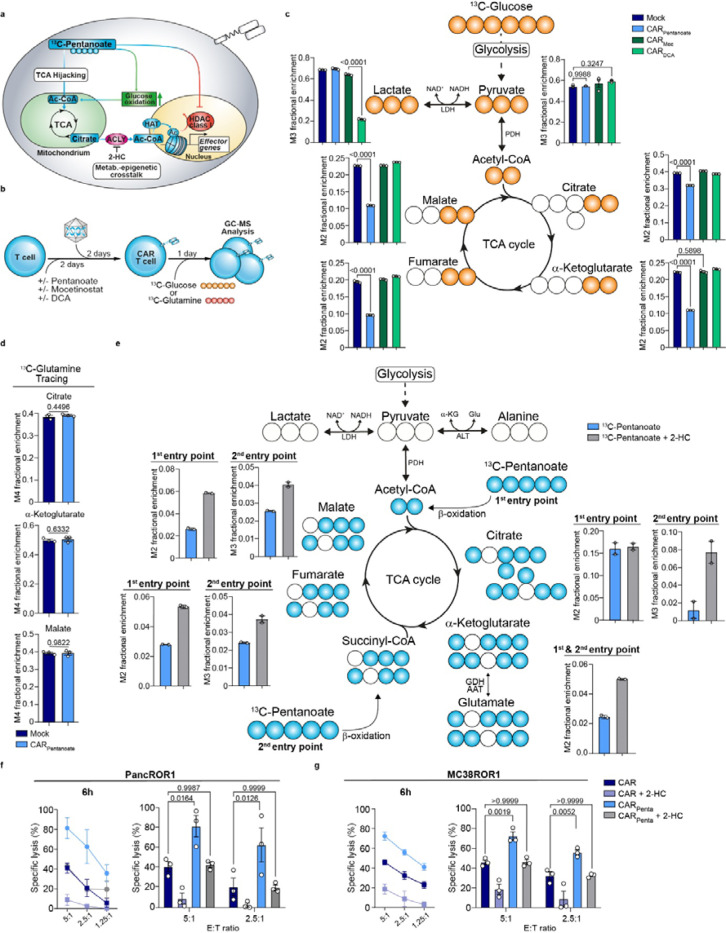
Pentanoate feeds into the TCA and becomes part of the epigenetic imprint
While DCA and pentanoate both influence mitochondrial activity, they might differ in their effect on glycolytic and oxidative pathway intermediates (Fig. 5a). To trace the pathway intermediates, we pulsed our engineered CAR T cells with either 13C-labeled glucose or 13C - glutamine tracers, prior to analysis via GC-MS (Fig. 5b). Tracing of 13C-glucose revealed that less glucose was metabolized to lactate upon DCA treatment, confirming its activity as PDK inhibitor. No significant changes in pyruvate were observed for any of the small molecules used (Fig. 5c). Of note, we detected a strong decrease in glucose-derived citrate in CARPenta T cells that was not observed for mocetinostat or DCA (Fig. 5c). However, the impact on the glycolysis end product was rather minor with a reduction of approximately 25%. This trend was more pronounced with a reduction of approximately 50% glucose-derived labeling for downstream TCA intermediates such as α-ketoglutarate, fumarate and malate, as compared to control CAR T cells. In contrast, although DCA reduced cellular lactate levels, its effects on glucose oxidation did not change the overall glucose-dependent citrate production. These data highlight pentanoate’s capacity to reprogram the TCA in T cells which cannot be mimicked by conventional drugs.
Figure 5: Pentanoate feeds into the TCA and becomes part of the epigenetic imprint.
a, Scheme illustrating pentanoate modulation and metabolism hijacking of T cells. b, Schematic for the generation of CAR+ T cells in combination with pretreatment prior to metabolite tracing c, GC-MS isotope tracing of 13C-glucose-derived metabolites in CAR T cells engineered in the presence of specified substances. Histograms show fractional enrichment of 13C-glucose-derived TCA metabolites. Mean ± SEM from n = 3 biological replicates. d, GC-MS isotope tracing of 13C-labeled glutamine in untreated or pentanoate- treated CAR T cells regarding citrate, α-ketoglutarate and malate. Mean ± SEM from n = 3 biological replicates. e, GC-MS tracing of 13C-pentanoate in CD8+ CAR T cells pretreated with indicated substrates. Histograms show the frequency of pentanoate-derived carbons in the downstream TCA cycle intermediates. n=2 independent T cell pools. f and g, PancROR1 (f) or MC38ROR1 (g) tumor killing of CAR+ T at different E:T ratios after 6 h. Mean ± SEM from n = 3.
Statistical analysis was performed using unpaired two-tailed Student’s t test (d), one-way ANOVA (c) or two-way ANOVA with Tukey’s multiple-comparison test (f and g).
We next addressed which alternative source was used by CARPenta T cells to replenish the citrate carbons. Upon butyrate treatment, T cells may fuel the TCA via glutamine anaplerosis to uncouple it from glycolysis [20]. By contrast, pulsing of CARPenta T cells with 13C-glutamine did not show a preference for incorporation into the TCA intermediates (Fig. 5d). Hence, fatty acid oxidation and metabolization of pentanoate might contribute to citrate generation. To understand how pentanoate is metabolized, we next administered 13C-pentanoate during the CAR manufacturing. GC-MS analysis revealed the incorporation of pentanoate-derived carbons into citrate via the acetyl-CoA entry point. Additionally, the labeled carbons entered the TCA through a second entry point via the succinyl-CoA route (Fig. 5e). This is a unique feature of pentanoate attributed to the C5 aliphatic chain, which cannot be emulated by acetate, propionate or butyrate. The combined citrate amounts derived from both entry points correspond to the gap detected in the previous glucose tracing experiment which underlining a limited preference for pentanoate over glucose (Fig. 5c, e). Besides TCA intermediates being required for energy homeostasis, conversion of citrate into acetyl-CoA by the nuclear ATP-citrate lyase (ACLY) serves as a source for histone acetylation, thereby linking metabolism and epigenetic regulation. To test whether ACLY directs the pentanoate-derived citrate flux, we inhibited ACLY via 2-hydroxy citrate (2-HC) in the presence of the 13C-pentanoate tracer (Fig. 5e). Interestingly, 2-HC supplementation during CARPenta T cell manufacturing not only led to a 2-fold increase in pentanoate labeling of glutamate, fumarate and malate downstream of citrate from the first entry point, but to an additional 4-fold increase of pentanoate-derived citrate from the second one. This suggests that the latter might be used for histone acetylation in the nucleus and could be accumulated upon ACLY blockade in the TCA. To investigate the functional outcome of this crosstalk, we evaluated CARPenta T and control CAR T cells that were engineered in the absence and presence of 2-HC in killing assays. Specific lysis and antigen-specific cytokine analysis showed that 2-HC administration reduced the functionality of CARPenta T cell significantly emphasizing the importance of the epigenetic-metabolic crosstalk engaged by pentanoate (Fig. 5f, g and Extended Data Fig. 13a-c).
Epigenetic-metabolic rewiring enhances CAR T cell efficacy, persistence and resistance to TME factors
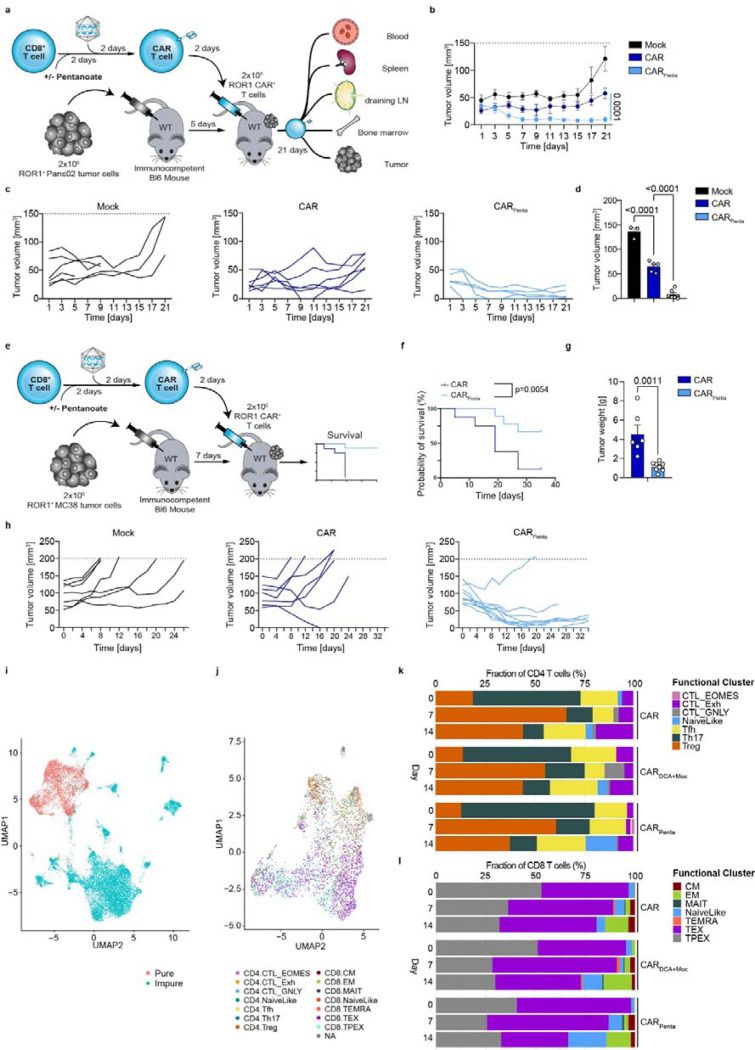
To assess the therapeutic potential of CARPenta T cells, we established syngeneic tumor models in fully immunocompetent mice, accounting for hostile TME conditions that are lacking in conventional xenogeneic NSG models. CARPenta T cells were transferred into mice bearing 5-day-old PancROR1 tumors, prior to endpoint analysis on day 21 post transfer (Fig. 6a). The Panc02 model develops a cold, immunosuppressive and T cell-excluding TME. Treatment of mice with pentanoate-engineered ROR1 CAR T cells led to significantly improved tumor control, approaching mass clearance in some mice starting 5–7 days after administration. In contrast, the conventionally engineered T cell group showed inferior anti-tumor activity and signs of losing efficacy towards the end of the experiment (Fig. 6b, c). Analysis at the endpoint showed low residual tumor mass and an increase in both CARPenta T cells as well as endogenous CD8+ tumor infiltrating lymphocytes (TILs) (Fig. 6d and Extended Data Fig. 14a). Further, CARPenta T cells in the draining lymph node (dLN), spleen, bone marrow and blood were more persistent (Extended Data Fig. 14b). Intracellular cytokine staining revealed elevated frequencies of TNF-α + IFN-γ+ and Granzyme B + IL-2 + CAR T cells in the spleen and to a lesserextent in the dLN. Significant differences were found in the tumor which reflected preservation of the cytokine profile in vitro (Fig. 2h, i and Extended Data Fig. 14c-e). Interestingly, in vivo transfer of a polyclonal T cell population similarly showed an antigen-independent expansion and persistence in the periphery upon pentanoate but not butyrate treatment (Extended Data Fig. 15). By contrast, pentanoate treatment for 3 days after CAR engineering limited persistence upon in vivo transfer into immunocompetent hosts (Extended Data Fig. 16a-c). This emphasizes the importance of timing the modulation within an early activation window when chromatin is primed to adopt an open state.
Figure 6: Epigenetic-metabolic rewiring enhances efficacy and persistence in solid malignancies.
a, Scheme illustrating the analysis of ROR1 CAR T cells in a PancROR1 tumor model. b, PancROR1 tumor growth after treatment with CD8+ CAR T cells. c, Tumor volume for mock, CAR T cells or CARPenta T cells over the experimental course. d, Tumor volume at endpoint day 21. Mean ± SEM from n = 5 mice/group for mock, n = 6 mice/group for CAR treated groups. e, Scheme illustrating the analysis of ROR1 CAR T cells in a MC38ROR1 tumor model. f, Survival of MC38ROR1 tumor bearing mice. Mean ± SEM from n = 8 mice for control group, n = 9 for CAR or CARPenta groups. g, Tumor weight at the endpoint. Mean ± SEM from n = 6 for CAR or CARPenta groups. h, Single tumor volume over the course of the experiment. i, Uniform Manifold Approximation and Projection (UMAP) showing the distribution of immune cells collected from mice using scRNAseq data in the LSI space. Each point represents one cell. Data points are color-coded based on cell annotations. Red indicates pure (T cells), blue impure (non-T cells). j, UMAP showing the distribution of T cells based on their cell states. Data points are color-coded based on their cluster affiliation. k and l, CD4 (k) and CD8 (l) T cell population changes following different treatments.
Statistical analysis was performed using one-way ANOVA with Tukey’s multiple-comparison test (b, d) and unpaired two-tailed Student’s t test (e-h and k). Survival curves were compared by the log-rank Mantel-Cox test (j).
We next assessed the impact of CARPenta T cells on mouse survival in an aggressive MC38ROR1 model with hot TME characteristics (Fig. 6e). Monitoring over 34 days post transfer revealed superior tumor control and survival as compared to the control group with conventionally engineered CAR T cells (Fig. 6f-h). While the control group delayed tumor growth before the experimental cut-off size was reached, pentanoate-engineered CAR T cells showed homogeneous, synchronized response and tumor control. Two mice were taken out of the experiment due to skin irritation at the tumor site.
To better understand at transcriptomic level the effects of the drug treatments on the immune microenvironment, we performed single cell RNA sequencing (scRNA-seq). Ex vivo isolated immune cells were obtained from tumor-bearing mice treated with CAR, CARDCA+Moc and CARPenta T cells on day 7 and day 14 post transfer, respectively, and the pre-infusion products prior to scRNA-seq (n=24, n=4 mice /treatment/ point in time, Extended Data Fig. 17a). After performing the QC steps a total of 16908 cells were identified. The UMAP representing the distribution of the cells based on conditions, time points and mouse of origin are presented in Extended Data Fig. 17b-d. By using scGate we were able to annotate these cells and identify 4774 of pure T cells that were further divided in 14 clusters based on their cell state (Figure 6i, j and Extended Data Fig. 17e). Of interest, following treatment with pentanoate we observed an enrichment of CD4 and CD8 naïve-like T cells (TCF7+, CCR7+, PD1-, TOX-) while a decrease in CD8 TEX (GZMK-, TCF7-, TOX+, HAVCR2+, PD1+) was noted (Figure 6k, l). Taken together, these data provide evidence for pentanoate’s ability to increase the fitness of T cells creating a more active immune microenvironment that supports T cell anti-tumor activity and reduce T cell exhaustion.
Conceivably, pentanoate-mediated epigenetic-metabolic reprogramming might also allow for enhanced resistance to hostile TME factors present in the solid tumors. Thus, we challenged CAR and CARPenta T cells for tumor elimination under hypoxic conditions. CARPenta T cells sustained their superior lytic activity, suggesting better maintenance of effector functions despite the oxygen-reduced environment (Extended Data Fig. 18a). We next engineered CAR T cells under low IL-2 conditions mimicking the IL-2-depleted environment characteristic of Treg-rich TMEs. Analysis of tumor cell survival showed that these under conditions, typically unfavorable to T cell functionality, CARPenta T cells retained superior lytic activity, potentially due their ability to produce autocrine IL-2 (Fig. 2l, m and Extended Data Fig. 18b).
Our results demonstrate that introducing the commensal metabolite pentanoate into CAR manufacturing confers long-lasting improvements in effector function, in vivo persistence and resistance to immunosuppressive factors in hot and cold TMEs, generating enhanced cell products with improved activity against hard-to-treat malignancies.
Discussion
Both homeostatic regulation and pathophysiologic development have been linked to the composition of the gut microbiome. The direct interaction between host and commensals, as well as soluble metabolites reaching distant sites in the body, are major influences on the immune system which can be therapeutically exploited. Here we demonstrate that the microbial metabolite pentanoate is predictive for CAR T cell patient survival and can be integrated into CAR T cell engineering to exploit the benefits of the host-microbiome interaction.
Previous reports in the context of ICI have revealed that commensals such as Bifidobacterium and Akkermansia muciniphila enhance anti-tumor immunity in mice in a T cell-dependent manner and are associated with better patient prognosis [6, 7, 9]. In line, correlations between antibiotics treatment and worse patient outcomes were recently established for CAR T cell therapy (Fig. 1a, b) [10, 9]. Initial attempts to exploit microbiome-based approaches for cancer immunotherapy involved fecal microbiota transplantations from responders into ICI-resistant patients after microbiome reset as well as patients with severe gastrointestinal graft-versus-host disease (GVHD) following allogeneic stem cell transplantation (allo-SCT) [21, 22]. A smaller fraction of the ICI-treated patients indeed achieved a response, but the remainder of the cohort remained resistant. Likewise, the overall response rates of GVHD patients were between 30 and 50% [23].
Furthermore, contradictory findings have slowed the translation of other microbiome-based therapies. Similar to our findings in CAR T patients, detection of high levels of the SCFAs propionate and butyrate was correlated with decreased PFS in ICI patients (Fig. 1d) [12]. The same class of metabolites was initially described as potent gatekeeper of intestinal homeostasis capable of inducing Treg differentiation and repressing allergic, inflammatory and autoimmune reactions [16, 13, 11, 24]. By contrast, we have demonstrated that SCFAs induce not only regulatory T and B cells but also CTL-associated factors in CD8 T cells (Fig. 2) [11]. This emphasizes cell type- and context-dependent effects of commensal metabolites rather than a broadly applicable phenotypic change [24, 14, 25, 20]. As many modes of action shaping immunosuppressive and –stimulatory phenotypes overlap, an in-depth mechanistic understanding of each commensal metabolite is required to unlock their full potential for the development of new therapeutic approaches such as CAR T cell therapy. Here, we show that among all SCFAs, pentanoate is the only one predictive of patient survival in a three-centric CAR T cell cohort (Fig. 1d-g). Despite its HDAC-inhibitory function, which is a shared feature among SCFAs, pentanoate did not induce Treg differentiation in CD4 T cells in contrast to butyrate and propionate (Extended Data Fig. 4a, b). Notably, the presence of CAR Tregs in infusion products can have detrimental effects in respect to relapse in patients [17]. In line with our in vitro findings, a small reduction of Tregs in the pre-infusion product (D0) and among the TILs (D14) was observed in the CARPenta T cell group (Fig. 6k). Thus, the choice of an HDAC inhibitor must be carefully made based on its interaction with the raw material when engineering immune cells. Furthermore, the importance of epigenetic regulation has been highlighted by the generation of mice bearing HDAC class I-deficient T cells whose CD4 T cells commit to the CD8 lineage during aging [26]. Additionally, other epigenetic modifications such as methylation were shown to be factors determining the fate and function of CAR T cell products [27, 28].
Besides being natural HDAC inhibitors, SCFAs modulate cellular metabolism. The link between metabolic fitness and potency of cell products has prompted approaches to actively shape metabolic pathways by overexpression of nutritional transporters or master regulators like PGC-1α [29 32]. In particular, mitochondrial metabolism has been identified as a critical factor in T cell memory formation [29, 33, 20, 34]. The choice of the CAR costimulatory domain was suggested to affect efficacy and persistence of the engineered T cell via balancing of glycolytic and mitochondrial function [35, 19]. In agreement with this idea, pharmacologic inhibition of the mitochondrial pyruvate carrier in CAR T cells results in improved anti-tumor response [33]. However, we demonstrated that DCA did not improve CAR T cell efficacy to the same extent as pentanoate, despite engaging similar pathways. In contrast to clinically available drugs, pentanoate hijacked the TCA through two different entry points post β-oxidation, which is a unique feature of the odd-numbered C5 aliphatic chain. Of note, Bachem and colleagues reported that pentanoate’s structurally closest relative butyrate elicits its metabolic function primarily via glutamine anaplerosis [20].
The enzyme ACLY links metabolic output and histone acetylation in mammalian cells [36]. Its inhibition via 2-HC in pentanoate-engineered CAR T cells led to accumulation of SCFA-derived citrate in the TCA, primarily from the succinyl-CoA entry route, and consequently a decrease of cytotoxic function (Fig. 5e-g). These findings suggest that pentanoate-derived carbons contribute to histone acetylation and differentiation towards an effector phenotype.
Strikingly, superior anti-tumor activity was achieved upon a single treatment with pentanoate and retained after removal, both in vitro and in vivo. CARPenta T cell administration resulted in enhanced tumor control compared to conventionally engineered T cells in two different syngeneic, immunocompetent solid tumor models with cold and hot TME, respectively (Fig. 6a-h). Short-term pentanoate treatment led to enhanced expansion and persistence in the periphery, greater tumor infiltration and a naive-like differentiation with a decrease in exhaustion (Fig. 6j-l). By contrast, the effect of butyrate on T cells in the context of infection was previously reported as inducing a memory phenotype [20]. Pentanoate-engineered CAR T cells maintained improved lytic activity under hypoxia and low IL-2 conditions suggesting resistance against TME-mediated dysfunction (Extended Data Fig. 18). Moreover, the enhanced autocrine IL-2 secretion may facilitate competitive fitness over endogenous immune cells, even in absence of lymphodepletion. While administration of lymphodepleting drugs is typically a prerequisite to generate a niche for CAR T cells, it renders patients susceptible to infections and necessitates preventive antibiotic treatment. This countermeasure in turn can affect pentanoate abundance and survival upon CAR T cell treatment (Fig. 1a-d) [10, 9]. Thus, approaches requiring minimal lymphodepletion with retained immune cell activity would be favorable for clinical application.
Although previous studies highlighted the relevance and potential of microbiome modulation in influencing immunotherapy responses, the clinical implementation is only slowly picking up pace. Designer consortia, pre- and antibiotics or phages face challenges as the dynamics of commensal colonization and the diversity of host immune factors might complicate their application [37, 8, 25]. The use of probiotic strains bears the risk for translocation and infections. Our study introduces postbiotic pentanoate as a promising model of the microbiome-host relationship, highlighting its advantages by incorporating epigenetic and metabolic adjustments into CAR T cell products, independently of gene editing, CAR target and co-stimulatory domain (Fig. 2b-m and Extended Data Fig. 10a, b). This approach enables a safer and more manageable side effect profile while reducing the complexity and genotoxicity associated with multi-factor genetic engineering. The integration of microbial metabolites in clinical-grade GMP manufacturing processes can offer prolonged modulation, improved metabolic fitness and standardized medicine development. Pentanoate’s unique characteristics and predictive potential highlight the microbiome as a source to mine for novel physiological drugs and biomarkers that can be exploited in tailored immunotherapy settings.
Methods
Study design and patients
The study was designed as a prospective observational study. All patients receiving CAR T cell therapy were eligible for this study regardless of the target antigen. Participants were enrolled from 05/2020 to 12/2023 at two centers: the University Medical Center Regensburg and the University Hospital Heidelberg. All patients provided informed written consent. The study was approved by the Institutional Ethics Review Board of the University of Regensburg, vote no. 21–2521-101. Patient characteristics are summarized in Suppl. Table 1. Fecal samples were collected from all patients prior to CAR T cell infusion. Fecal samples were prepared and short-chain fatty acids were measured as previously described [38]. Briefly, metabolites from 100mg feces were extracted using a 15-ml bead-beater tube (CKMix50, 15 ml, Bertin Technologies) filled with 2.8-mm and 5.0-mm ceramic beads and a bead-beater (Precellys Evolution, Bertin Technologies) at 10,000 rpm (3 rounds of 30s with 15s breaks). Methanol-based dehydrocholic acid extraction solvent (5 ml, c = 1.3 μmol l−1) was added as an internal standard to account for work-up losses. Short chain fatty acids were measured using the 3-NPH method in a QTRAP 5500 triple quadrupole mass spectrometer (SCIEX) coupled to an ExionLC AD (SCIEX) ultra-highperformance liquid chromatography system. Data were analyzed with MultiQuant 3.0.3 (SCIEX) and MetaboAnalyst. Stool samples have limited availability due to the individual amount obtained from each patient.
An external cohort of 60 patients with large B cell lymphoma or follicular lymphoma, treated with commercial CD19-directed CAR-T cells, was used to validate the association between stool pentanoate levels and CAR T therapy outcomes (Suppl. Table 2). Stool samples collected between day −30 and day 0 (median: day −3 [IQR: −4.3 to −1]) were aliquoted and stored at −80°C. One sample per patient was sent to Metabolon for metabolomic profiling
Sample Preparation:
Feces samples (fresh/frozen) are analyzed for eight short chain fatty acids: acetic acid (C2), propionic acid (C3), isobutyric acid (C4), butyric acid (C4), 2-methyl-butyric acid (C5), isovaleric acid (C5), valeric acid (C5) and caproic acid (hexanoic acid, C6) by LC-MS/MS. Samples are spiked with stable labelled internal standards and are homogenized and subjected to protein precipitation with an organic solvent. After centrifugation, an aliquot of the supernatant is derivatized. The reaction mixture is diluted, and an aliquot is injected onto an Agilent 1290 Infinity or Infinity II/ Sciex QTrap 5500 or 6500 LC MS/MS system equipped with a C18 reversed phase UHPLC column. The mass spectrometer is operated in negative mode using electrospray ionization (ESI).
Sample Analysis:
The peak area of the individual analyte product ions is measured against the peak area of the product ions of the corresponding internal standards. Quantitation is performed using a weighted linear least squares regression analysis generated from fortified calibration standards prepared immediately prior to each run. LC-MS/MS raw data are collected and processed using AB SCIEX software Analyst 1.6.3 and processed using SCIEX OS-MQ software
QA/QC:
Three levels of QCs are prepared in feces by diluting and/or spiking with stock solutions to obtain the appropriate concentrations for each level (low/med/high). Accuracy will be evaluated using the corresponding QC replicates in the sample runs. Targeted acceptance criteria are at least 50% of QC samples at each concentration level per analyte should be within ±20.0% of a set mean and at least 2/3 of all QC samples per analyte should fall within ±20.0% of the corresponding mean.
Animals
C57BL/6 wild-type mice were purchased from Charles River and maintained under specific pathogen free conditions at the Center for Experimental Medicine (ZEMM) at the University of Würzburg. Tbx21−/− mice (on C57BL/6 background) were maintained under specific pathogen free (SPF) conditions at the animal facility of the Philipps-University of Marburg, Germany. Mice were kept under a 12/12 h light/dark cycle between 20–24 °C in individually ventilated cages. Mice had access to standard chow and autoclaved water ad libitum and the health status of the animals was inspected by responsible animal caretakers. Male and female mice between 8–12 weeks old at the time of the experiment were used in this study. All animal protocols were approved by government (Approval number: 1457, Regierung von Unterfranken, Bayern, Germany).
Hdac1fl/flHdac2fl/fl (HDAC1–2cKO) CD4-Cre mice (Mouse Genome Informatics [MGI] 4440556 for Hdac1; MGI 4440560 for Hdac2) were previously described and kept under specific pathogen free conditions at the Medical University of Vienna [26, 39].
Cell lines
The mouse MC38 colon adenocarcinoma cell line was provided by the lab of Tobias Bopp/Toska Bohn. The mouse Panc02 OVA pancreatic tumor cell line was gifted from the Christian Bauer lab. All solid tumor cell lines were cultured in standard DMEM (Gibco) supplemented with 10 % heat-inactivated FCS (Gibco) and 1 % penicillin/streptomycin (Gibco) at 37 °C with 5 % CO2.
The mouse lymphoma cell line E -myc was supplied by the lab of Dirk Busch and cultured in standard RPMI 1640 (Gibco) with heat-inactivated 10 % FCS (Gibco) and 1 % penicillin/streptomycin (Gibco) at 37 °C with 5 % CO2.
The Platinum-E retroviral packaging cell line (Cell Biolabs) was cultured in standard DMEM (Gibco) supplemented with 10 % heat-inactivated FCS and 1 % penicillin/streptomycin (both Gibco) at 37 °C with 5 % CO2. Following thawing, PlatinumE cells were selected for 5 days using 1 μg/ml puromycin and 10 μg/ml blasticidin S (both Invivogen) and subsequently cultured. All cell lines were tested for mycoplasma contamination.
Virus production
Platinum-E cells for retroviral packaging were co-transfected with the desired construct in addition with retroviral packaging construct pCL-10A1, using the Effectene transfection reagent (QIAGEN) according to the manufacturer’s instructions. The retroviral supernatant was collected 48 and 72 h after transfection, pooled and stored at −80 °C until use.
In vitro T cell differentiation and culture
For in vitro cultures, murine T cells from male or female mice were isolated from single cell suspension of spleen and lymph nodes. Isolation of CD8+ T cells was performed by positive selection (Miltenyi Biotech), followed by a negative selection for CD4+ T cells (Invitrogen). T cells were cultured unless stated otherwise in modified RPMI 1640 medium (Gibco) supplemented with 10 % heat-inactivated FCS (Gibco), 50 μM 2-mercaptoethanol (Gibco), 1 % penicillin/streptomycin and 1 % GlutaMAX-I (Gibco) at 37 °C with 5 % CO2.
Culture plates were pre-coated with 10 μg/ml polyclonal anti-hamster IgG (MP Biomedicals) for 2 h and washed once with PBS (Gibco). For Th1 differentiation, 0.5×106 CD4+ T cells were activated in the presence of 1 μg/ml anti-CD3 (Biolegend, 145–2C11), 1 μg/ml anti-CD28 (Biolegend, 37.51), 1 μg/ml anti-IL-4 (Biolegend, BVD4–1D11), 50 U/ml recombinant human (rh) IL-2 (Miltenyi Biotech) and 10 ng/ml IL-12 (Miltenyi Biotech).
For supoptimal CTL differentiation, 0.5×106 CD8+ T cells were cultured with 1 μg/ml anti-CD3, 1 μg/ml anti-CD28, 1 μg/ml anti- IFN-γ (Invitrogen, XMG1.2), 1 μg/ml anti-IL-4 and 50 U/ml rh IL-2.
For iTreg differentiation, CD4+ T cells were activated with 1 μg/ml anti-CD3, 0.5 μg/ml anti-CD28, 100U/ml rh IL-2, 2 μg/ml anti- IFN-γ, anti- IL-4 and 2 ng/ml rhTGF-β1 (R&D Systems).
In some experiments, medium of cells was added with 2 mM sodium Pentanoate (Ambeed), 5 mM dichloroacetate (Merck), 100 nM mocetinostat (Biomol), 1 μM TMP-195 (Biomol), 1 mM Butyrate (Sigma), 2 mM Propionate (Merck), 10 mM Acetate (Merck) or a combination of DCA and mocetinostat once 2 hours after activation. Furthermore, 5 mM 2-hydrocycitrate (2-HC) (Sigma) was added to cell culture at indicated time points. All exogenous metabolites were dissolved in DMSO solutions according to the manufacturer’s recommendation.
Generation of murine CAR T cells
48 hours after activation, the medium of the cells was removed and stored at 4°C. Cells were transduced with retroviral supernatant in addition with 10 μg/ml polybrene by spin-infection at 800 rpm for 2 hours at 32 °C and placed in an incubator for 4 hours. Afterwards, the supernatant was replaced by the stored medium. The next day, T cells were put into new culture-plates. On day 4, fresh medium including 50 U/ml rh IL-2, 10 ng/ml IL-7 and 10 ng/ml IL-15 (all Miltenyi) was added. Cells were maintained between 0.5×106 and 2×106 cells/ml and expanded until day 7.
Flow cytometry
T cells were harvested, washed once with PBS and stained with live/dead marker (Biolegend) for 20 min at RT. After another washing step, surface antigens with fluorophore-conjugated antibodies were stained for 20 min in PBS at 4 °C in the dark. For staining of intracellular cytokines, cells were stimulated with 100 ng/ml phorbol-12-myristat-acetate (PMA) and 1 μg/ml Ionomycin in the presence of 5 μg/ml brefeldin A for 4–5 hours at 37 °C. Cells were fixed with 2 % paraformaldehyde (Invitrogen) for 20 min at 4°C, before being stained for intracellular cytokines with antibodies in Saponinbuffer for 30 min at 4 °C.
For the detection of transcription factors and histone modifications, activated T cells were fixed with the Foxp3 / Transcription Factor Staining buffer set following manufacturer’s instructions (eBioscience) and staining with described antibodies was conducted for 30 min in Saponinbuffer at 4 °C. To assess neutral lipid content, cells were stained with BODIPY 493/503 (Invitrogen) for 20 min at RT. For the quantification of mitochondrial volume, T cells were labeled with 200 nM MitoTracker Deep Red (Invitrogen) and stained for 30 min at 37 °C. To measure glucose uptake, cells were incubated with fluorescent glucose analog 2-NBDG (Cayman Chemicals) for 20 min at 37 °C. To assess phosphorylation of intracellular S6 cells were stained as described previously [15].
All data was collected on a BD Canto II and analyzed using FlowJo. MFI marks median fluorescence intensity calculated by FlowJo. For the list of antibodies used for flow cytometry see Suppl. Table 2. Examples of the gating strategy are shown in Extended Data Fig. 18.
Cytotoxicity assay
To determine antigen-specific tumor cell lysis, 5 × 103 ffLuc-transduced tumor cells were co-cultured with CAR-transduced or untransduced (UTD) T cells at effector:target (E:T)-ratios of 10:1, 5:1, 2.5:1 or 1.25:1 in triplicates in RPMI medium supplemented with 150 ng/ml D-Luciferin. For experiments under hypoxia conditions, cells were plated in an incubator with 2 % O2. The bioluminescence signal was measured on a Tecan Infinite 200 PRO plate reader after 4, 6, 8 and 24 h. Specific lysis was determined in reference to the corresponding untransduced T cells.
Cytokine secretion
Secretion of cytokines for murine CAR T cells was detected by ELISA kits for mouse IFN-γ, IL-2, Granzyme B (all Biolegend) and TNF-α (Invitrogen) according to the manufacturerer’s instructions. Cells were co-incubated with 5:1 or 2.5:1 E:T ratio with antigen-presenting tumor cells in triplicates for 24 h. Absorbance was measured using a Tecan Infinite 200 PRO plate reader.
HDAC docking methods
Structure predictions of HDACs 1–5, 7–9 with zinc ions (one zinc for HDACs 1 and 2, two zinc ions for HDACs 3–5 and 7–9) with and without palmitate were performed using AlphaFold 3 [40]. SCFAs and mocetinostat were docked into AlphaFold 3-generated HDAC1 models using the Swissdock attracting cavities docking engine [41–43]. Medium sampling exhaustivity and buried cavity prioritization parameters were used for all docking jobs. Random initial conditions (RIC) were set to 3 for docking SCFAs with HDAC1. RIC were set to 1 for docking mocetinostat into HDAC1 and pentanoate into HDAC4. Models were visualised and analysed using UCSF ChimeraX [44]. Palmitate molecules in the AlphaFold 3 models were truncated to C5 using UCSF ChimeraX and used for free energy calculations alongside models with palmitate. Free energy calculations were performed using PRODIGY-LIGAND [45, 46].
HDAC activity assays
For the impact of SCFAs on specific HDAC isoforms, the fluorogenic assay for each HDAC enzyme (HDAC1–3, HDAC5) was used (BPS Bioscience). Assays were conducted according to the manufacturer’s instructions, with measurements taken in triplicates using the FLUOstar Omega plate reader.
Metabolomic profiling
To analyze polar intracellular metabolites, CD8+ and CD4+ CAR T cells were generated as described above. After removal from the antibody on day 3, cells were put into medium containing 1 g/l 13C- glucose/ 13C- glutamine for 24 hours. Per condition, 1×106 T cells were washed with 0.9 % Natriumchloride. Metabolite extraction was conducted by incubation with ice-cold 80 % methanol including internal standards. Following an incubation time of 20 min at 4 °C, cells were pelleted and the supernatant containing the polar metabolites was transferred into a new tube and stored at −80 °C until further processing. For the tracking of pentanoate, 2 mM of 13C-labeled pentanoate was added at day 0 following activation and samples were taken from day 1 until day 4 after activation and isolated for metabolites as described above.
Metabolites were automatically derivatized using a Gerstel MPS. Derivatization was done with 15 μl of 2 % (w/v) methoxyamine hyprochloride (Thermo Scientific) in pyridine and 15 μl N-tertbutyldimethylsilyl-N-methyltrifluoroacetamide with 1 % tert-butyldimethylchlorosilane (Regis Technologies). Measurement was carried out by GC/MS with a 30 m DB-35MS + 5 m Duraguard capillary column (0.25 mm inner diameter, 0.25 μm film thickness) equipped in an Agilent 7890B gas chromatograph (GC) connected to an Agilent 5977A mass spectrometer (MS). The GC oven temperature was held at 80 °C for 6 min and steadily adjusted at 6 °C per min until reaching 280 °C where the temperature was held for 10 min. The quadropole was set to 150 °C. The MS source operated under electron impact ionization mode at 70 eV and was held at 230°C. Targeted single ion chromatogram measurements were conducted for pyruvate (174, 175, 176, 177, 178, 179; 10 scans per second), lactate (261, 262, 263, 264, 265, 266, 267; 10 scans per second), citrate (591, 592, 593, 594, 595, 596, 597, 598, 599, 600; 10 scans per second), α-ketoglutarate (346, 347, 348, 349, 350, 351, 352, 353, 354; 10 scans per second), fumarate (287, 288, 289, 290, 291, 292, 293; 10 scans per second), glutamate (432, 433, 434, 435, 436, 437, 438, 439, 440; 10 scans per second) and malate (419, 420, 421, 422, 423, 424, 425, 426; 10 scans per second).
All chromatograms were subsequently analyzed with the MetaboliteDetector software [47].
Preparation of cells for RNA-single cell sequencing
Cells of draining lymph nodes (dLN) and tumor-infiltrating lymphocytes (TILS) were isolated from tumor-bearing mice at day 7 and 14 post T cell infusion. Following incubation with TruStain FcX™ PLUS (anti-mouse CD16/32) antibody (Biolegend) to prevent unspecific binding, cells were stained with 2 μl of TotalSeqTM-C hashing antibodies (Biolegend) in wash buffer for 30 min at 4 °C.
Afterwards, 6 samples were pooled in one and suspended in 65 μl of PBS/0.04 % BSA.
Single cell RNA-sequencing
Chromium™ X/iX Controller was used for partitioning single cells into nanoliter-scale Gel Bead-In-EMulsions (GEMs) and Chromium GEM-X Single Cell 5’ Kit v3 kits for reverse transcription, cDNA amplification and library construction for all gene expression, TCR and hashtag libraries (10x Genomics), following manufacturer’s instructions. A SimpliAmp Thermal Cycler was used for amplification and incubation steps (Applied Biosystems). Libraries were quantified by a QubitTM 3.0 fluorometer (Thermo Fisher Scientific) and quality was checked using a 2100 Bioanalyser with High Sensitivity DNA kit (Agilent). Libraries were pooled and sequenced using the NextSeq 2000 platform (Illumina) in pairedend mode for gene expression as well as for the T-cell receptor repertoire and hashtags. Demultiplexed FASTQ files were generated with bcl-convert v4.0.3 (Illumina). Data were analyzed using the Cell Ranger 7.2.0 software suite pipelines available on the 10xGenomics website.
The Cell Ranger output of each pool (P1 to P9) was converted to a Seurat object using the Seurat package (Read10X and CreateSeuratObject). After adding the HTO data as an independent assay, cells were demultiplexed using the HTODemux function with kmeans clustering and positive.quantile =0.99. Only singlets were selected for further analysis. The percentage of mitochondrial genes (MT-) was determined for each cell using the PercentageFeatureSet function. We filtered out low quality cells based on the number of features (< 200 or > 6000) and the percentage of mitochondrial genes (> 20 %) using the Subset function. The 9 Seurat objects were merged into a single object regrouping a total of 16908cells. Normalization and integration were performed using NormalizeData and STACAS. RunPCA, runUMAP, FindNeighbors and FindClusters with 30dimensions were used to identify 21clusters of T cells. T cells were then annotated using scGate with the pre-defined gating mouse model, and only pure T cells were subset and used for downstream analysis. A total of 4774 pure T cells were annotated for the different CD4 and CD8 T cell subsets using ProjecTILs and the human reference atlas to characterize cell states.
In vivo models
For all experiments, group sizes were determined based on experience by previously published models. Tumor-bearing mice were randomly assigned to receive CAR T cell infusions, to ensure similar tumor sizes across groups before treatment. Tumor engraftment and T cell infusions were performed by blinded technicians. Generated CAR T cells were not sorted but additional non-transduced cells were added, so that the same amount of total T cell count was applied to each animal.
Panc02 tumor models
For adoptive transfer experiments with Panc02 mROR1 tumor cells, mice were subcutaneously (s.c.) injected with 2×106 tumor cells. On day 5 after tumor injection, 2×106 CD8+ CAR T cells were transferred intraperitoneally (i.p) into Bl6 mice.
For the analysis of transferred CAR T cells by scRNA sequencing, mice were subcutaneously (s.c.) injected with 1×106 tumor cells. After an inoculation time of 7 days, 1×106 CAR T cells with a 1:1 mixture of CD4:CD8 CAR T cells were transferred into tumor bearing mice. On day 7 and day 14 after T cell injection, 5 mice per group were sacrificed and samples of dLN and TILs were sent for RNA single cells sequencing.
MC38 tumor model
For adoptive transfer experiments with MC38 mROR1 tumor cells, mice were subcutaneously (s.c.) injected with 2×106 tumor cells in PBS with matrigel. Mice were treated with 2×106 CD8+ CAR T cells on day 7.
For all in vivo experiments, tumor progression was monitored by caliper every second day. Mice were monitored daily and euthanized when mice reached limits designated in the approved protocols including maximum tumor burden. At the endpoint of the experiments, peripheral blood was obtained by tail vein puncture. Cell pellets were resuspended in ACK lysis (Thermo Fisher) buffer for 10 min. Spleen, bone marrow, draining lymph node and tumors were collected. After determination of the weight, tumors were dissociated using the tumor dissociation kit (Miltenyi Biotech) and a GentleMACSTM Dissociator (Miltenyi Biotech) according to manufacturer’s constructions. All organs were passed through a 70 μM strainer to acquire single-cell suspensions. Tumor-infiltrating lymphocytes were isolated using CD45 (TIL) microbeads (Miltenyi Biotech). Consequent cell suspensions were washed with PBS and stained with live/dead marker (Biolegend) for 20 min at RT. After washing, samples were incubated with TruStain FcX™ (anti-mouse CD16/32) Antibody (Biolegend) to prevent unspecific binding for 15min at RT and then stained with FACS antibodies for analysis (List of antibodies in Suppl. Table 3).
Quantification and statistical analysis
The results are shown as mean ± standard error of the means (SEM). To determine the statistical significance of the differences between two experimental groups unpaired Student’s t tests were performed using Prism 9 software (GraphPad). For identify the statistical significance of the difference between more than to experimental groups one-way or two-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test were performed using Prism 9 software (GraphPad). For in vivo tumor growth curves, significance was determined at indicated timepoints on the blot by unpaired Student’s t-test comparing control group with the treatment group. Significance for the survival data was calculated using the log-rank Mantel-Cox test. Samples sizes were based on experience and complexcity of the experiment but no methods were used to determine normal distribution of the samples. Differences reached significance with p- values ≤0.05, p ≤ 0.0.1, p ≤ 0.001 or ≤ 0.0001. The figure legends contain the number of independent experiments or mice per group that were used in the respective experiment.
For the clinical study, statistical analyses were conducted using RStudio version 2023.12.1+402 (Posit, Boston, MA, USA) and R version 4.3.3 (The R foundation, Vienna, Austria). The level of significance was set at a two-sided p ≤ 0.05 with 95% confidence intervals. Grouped data are presented as violin plots. To compare two groups, based on the distribution of the data, the Wilcoxon-Mann-Whitney-Test (for non-normally distributed data) or the t-test (for normally distributed data) were conducted and adjusted for false discovery rate (FDR) using the rstatix package (version 0.7.2). Progression-free survival (PFS) was defined as the time from CAR T cell infusion to disease progression or death, whichever occurred first. Overall survival (OS) is defined as the timespan between the CAR T cell infusion and the patient’s death irrespective of the cause. Metabolite cutoffs were determined utilizing surv_cutpoint () function from the survminer package (version 0.4.9). Survival data are depicted as Kaplan-Meier curves. Differences between two groups were assessed with the log-rank test via the ggsurvfit () function and package (version 1.0.0). For comparisons of more than one group, a cox regression was performed using the coxph() function from the survival package (version 3.5–8).
Supplementary Material
Acknowledgments
The study was supported by the Austrian Science Fund project SFB F70 and by the Horizon 2020 Marie Slodowska Curie Innovative Training Network “Enlight-ten+”(Grant 955321 to WE), the Instituto de Salud Carlos III co-financed by European Regional Development Fund-FEDER “A way to make Europe” Red de Terapias Avanzadas TERAV (RD21/0017/0009 to FP, PMPTA22/00109 to JRR-M), Ministerio de Ciencia e Innovación co-financed by European Regional Development Fund-FEDER “A way to make Europe” (PID2022-137914OB-I00 to JRR-M), Centro de Investigación Biomédica en Red de Cáncer CIBERONC (CB16/12/00489 and CB16/12/00369 to FP), European Commission (H2020-JTI-IMI2-2019-18: Contract 945393; SC1-PM-08-2017: Contract 754658 to FP), the Engineered Air Chair in Cancer Research from University of Calgary (to PN), the European Molecular Biology Conference (EMBC) under EMBO Installation (Grant agreement number 5342-2023 to PP), the Ministry of Education, Science and Sports of Lithuania (Measure No. 12-001-01-01-01, Project No. S-A-UEI-23-10 to PP), the National Institutes of Health/National Cancer Institute (NIH/NCI) Memorial Sloan Kettering Cancer Center Support Grant (P30 CA008748 to RS), NIH-NCI K08-CA282987 (to RS) and the Research Council of Lithuania (LMTLT) (grant agreement number S-PD-24-74 to AS).
Moreover, support was received from the Innovative Medicines Initiative 2 Joint Undertaking, from the European Union’s Horizon 2020 research and innovation program and EFPIA (grant agreement No. 116026, T2EVOLVE to LW, DB, FP, MH and ML), the Wilhelm-Sander-Stiftung (Grant No. 2022.134.1 to AV, KM and ML), ERA-NET TRANSCAN-3 (EC co-funded call 2021, SmartCAR-T to KZM, PN, MH and ML), the Paula & Rodger Riney Foundation (to MvdB, MH and ML), IZKF Würzburg (S-511 to SS), the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG, SFB-1454 (project 432325352 to KH), SFB-1371 (Project 395357507 to HP and KK), TRR 221 (subproject A03 to MH, HE and ML; subproject A08 to HP and MP; and TRR338, (subproject A02; to MH, HE and ML, seed grant; to ML), the German Cancer Aid (70114547 to HP), the Bavarian Cancer Research Center (BZKF) (to MP, MAF, MH, ML and HP) and the ReforM program of the University Regensburg (to MP). This work was co-funded by the European Union (project MICROBOTS, Grant No. 101124680 to HP). We thank the Core Unit SysMed at the University of Würzburg for excellent technical support, scRNA-seq data generation and analysis. This work was supported by the IZKF at the University of Würzburg (project Z-6). We also thank all BayBioMS members for their help and fruitful discussions. The TripleTOF 6600 (Sciex) mass spectrometer and the Q Exactive HFX (Thermo) was funded in part by the German Research Foundation (INST 95/1434-1 FUGG and INST 95/1435-1 FUGG).
Competing interests
ML, MH and AV are listed as inventors on patent application WO2021/058811A1. MH is listed as an inventor on patent applications and granted patents related to CAR-T technologies that have been filed by the Fred Hutchinson Cancer Research Center, Seattle, WA and by the University of Würzburg, Würzburg, Germany. MH is a co-founder and equity owner of T-CURX GmbH, Würzburg, Germany. MH received honoraria from Celgene/BMS, Janssen, Kite/Gilead. MvdB has received research support and stock options from Seres Therapeutics and stock options from Notch Therapeutics and Pluto Therapeutics; he has received royalties from Wolters Kluwer; has consulted, received honorarium from or participated in advisory boards for Seres Therapeutics, Rheos Medicines, Ceramedix, Pluto Therapeutics, Thymofox, Garuda, Novartis (Spouse), Synthekine (Spouse), Beigene (Spouse), Kite (Spouse); he has IP Licensing with Seres Therapeutics and Juno Therapeutics; and holds a fiduciary role on the Foundation Board of DKMS (a nonprofit organization). PN received honoraria from BMS, Janssen, Sanofi and Pfizer as a consultant/advisory board member. MAF received honoraria from Novartis and Sanofi and travel grants from Sanofi. HP is a consultant for Gilead, Abbvie, Pfizer, Novartis, Servier, and Bristol Myers-Squibb. The remaining authors declare no financial conflict of interest.
Data availability
The authors declare that data supporting the findings of this study are available within the paper and its supplementary information files. Data derived from single cell sequencing experiments will be made publicly available upon manuscript acceptance.
References
- 1.Narayan V, Barber-Rotenberg JS, Jung I-Y et al. (2022) PSMA-targeting TGFβ-insensitive armored CAR T cells in metastatic castration-resistant prostate cancer: a phase 1 trial. Nat Med 28:724–734. 10.1038/s41591-022-01726-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Bruggen JAC, Martens AWJ, Fraietta JA et al. (2019) Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8+ T cells and impede CAR T-cell efficacy. Blood 134:44–58. 10.1182/blood.2018885863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kespohl M, Vachharajani N, Luu M et al. (2017) The Microbial Metabolite Butyrate Induces Expression of Th1-Associated Factors in CD4+ T Cells. Front Immunol 8. 10.3389/fimmu.2017.01036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith PM, Howitt MR, Panikov N et al. (2013) The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vétizou M, Pitt JM, Daillère R et al. (2015) Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350:1079–1084. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sivan A, Corrales L, Hubert N et al. (2015) Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350:1084–1089. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Routy B, Le Chatelier E, Derosa L et al. (2018) Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359:91–97. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 8.Tanoue T, Morita S, Plichta DR et al. (2019) A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565:600–605. 10.1038/s41586-019-0878-z [DOI] [PubMed] [Google Scholar]
- 9.Stein-Thoeringer CK, Saini NY, Zamir E et al. (2023) A non-antibiotic-disrupted gut microbiome is associated with clinical responses to CD19-CAR-T cell cancer immunotherapy. Nature Medicine:1–11. 10.1038/s41591-023-02234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith M, Dai A, Ghilardi G et al. (2022) Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat Med 28:713–723. 10.1038/s41591-022-01702-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luu M, Pautz S, Kohl V et al. (2019) The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun 10:760. 10.1038/s41467-019-08711-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coutzac C, Jouniaux J-M, Paci A et al. (2020) Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun 11:2168. 10.1038/s41467-020-16079-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arpaia N, Campbell C, Fan X et al. (2013) Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451–455. 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang PV, Hao L, Offermanns S et al. (2014) The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proceedings of the National Academy of Sciences 111:2247–2252. 10.1073/pnas.1322269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luu M, Riester Z, Baldrich A et al. (2021) Microbial short-chain fatty acids modulate CD8+ T cell responses and improve adoptive immunotherapy for cancer. Nat Commun 12:4077. 10.1038/s41467-021-24331-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith PM, Howitt MR, Panikov N et al. (2013) The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 341:569–573. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haradhvala NJ, Leick MB, Maurer K et al. (2022) Distinct cellular dynamics associated with response to CAR-T therapy for refractory B cell lymphoma. Nature Medicine 28:1848–1859. 10.1038/s41591-022-01959-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michelakis ED, Sutendra G, Dromparis P et al. (2010) Metabolic modulation of glioblastoma with dichloroacetate. Science Translational Medicine 2:31ra34. 10.1126/scitranslmed.3000677 [DOI] [PubMed] [Google Scholar]
- 19.Kawalekar OU, O’Connor RS, Fraietta JA et al. (2016) Distinct Signaling of Coreceptors Regulates Specific Metabolism Pathways and Impacts Memory Development in CAR T Cells. Immunity 44:380–390. 10.1016/j.immuni.2016.01.021 [DOI] [PubMed] [Google Scholar]
- 20.Bachem A, Makhlouf C, Binger KJ et al. (2019) Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8+ T Cells. Immunity 51:285–297.e5. 10.1016/j.immuni.2019.06.002 [DOI] [PubMed] [Google Scholar]
- 21.Routy B, Lenehan JG, Miller WH et al. (2023) Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat Med 29:2121–2132. 10.1038/s41591-023-02453-x [DOI] [PubMed] [Google Scholar]
- 22.Thiele Orberg E, Meedt E, Hiergeist A et al. (2022) Identification of Bacterial and Viral Consortia for the Production of Intestinal Microbiota-Derived Metabolites Associated with Protection in Allogeneic Stem Cell Transplantation Patients. Blood 140:7619–7621. 10.1182/blood-2022-157423 [DOI] [Google Scholar]
- 23.Weber D, Meedt E, Poeck H et al. (2024) Fecal Microbiota Transfer in Acute Graft-versus-Host Disease following Allogeneic Stem Cell Transplantation. Visceral Medicine:1–6. 10.1159/000538303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trompette A, Gollwitzer ES, Yadava K et al. (2014) Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature Medicine 20:159–166. 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- 25.Hu Y, Li J, Ni F et al. (2022) CAR-T cell therapy-related cytokine release syndrome and therapeutic response is modulated by the gut microbiome in hematologic malignancies. Nat Commun 13:5313. 10.1038/s41467-022-32960-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucheron N, Tschismarov R, Goeschl L et al. (2014) CD4(+) T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2. Nat Immunol 15:439–448. 10.1038/ni.2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain N, Zhao Z, Koche RP et al. (2024) Disruption of SUV39H1-Mediated H3K9 Methylation Sustains CAR T-cell Function. Cancer Discov 14:142–157. 10.1158/2159-8290.CD-22-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prinzing B, Zebley CC, Petersen CT et al. (2021) Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Science Translational Medicine 13:eabh0272. 10.1126/scitranslmed.abh0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lontos K, Wang Y, Joshi SK et al. (2023) Metabolic reprogramming via an engineered PGC-1α improves human chimeric antigen receptor T-cell therapy against solid tumors. Journal for ImmunoTherapy of Cancer 11. 10.1136/jitc-2022-006522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y, Kotchetkov IS, Dobrin A et al. (2024) GLUT1 overexpression enhances CAR T cell metabolic fitness and anti-tumor efficacy. Molecular therapy : the journal of the American Society of Gene Therapy 32:2393–2405. 10.1016/j.ymthe.2024.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panetti S, McJannett N, Fultang L et al. (2023) Engineering amino acid uptake or catabolism promotes CAR T-cell adaption to the tumor environment. Blood Advances 7:1754–1761. 10.1182/bloodadvances.2022008272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan JD, Scheffler CM, Munoz I et al. (2024) FOXO1 enhances CAR T cell stemness, metabolic fitness and efficacy. Nature 629:201–210. 10.1038/s41586-024-07242-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenes M, Jaccard A, Wyss T et al. (2022) The mitochondrial pyruvate carrier regulates memory T cell differentiation and antitumor function. Cell Metabolism 0. 10.1016/j.cmet.2022.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaccard A, Wyss T, Maldonado-Pérez N et al. (2023) Reductive carboxylation epigenetically instructs T cell differentiation. Nature 621:849–856. 10.1038/s41586-023-06546-y [DOI] [PubMed] [Google Scholar]
- 35.Zhao Z, Condomines M, van der Stegen SJC et al. (2015) Structural Design of Engineered Costimulation Determines Tumor Rejection Kinetics and Persistence of CAR T Cells. Cancer cell 28:415–428. 10.1016/j.ccell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wellen KE, Hatzivassiliou G, Sachdeva UM et al. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324:1076–1080. 10.1126/science.1164097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan Mirzaei M, Deng L (2022) New technologies for developing phage-based tools to manipulate the human microbiome. Trends in Microbiology 30:131–142. 10.1016/j.tim.2021.04.007 [DOI] [PubMed] [Google Scholar]
- 38.Thiele Orberg E, Meedt E, Hiergeist A et al. (2024) Bacteria and bacteriophage consortia are associated with protective intestinal metabolites in patients receiving stem cell transplantation. Nat Cancer 5:187–208. 10.1038/s43018-023-00669-x [DOI] [PubMed] [Google Scholar]
- 39.Preglej T, Hamminger P, Luu M et al. (2020) Histone deacetylases 1 and 2 restrain CD4+ cytotoxic T lymphocyte differentiation. JCI Insight 5. 10.1172/jci.insight.133393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abramson J, Adler J, Dunger J et al. (2024) Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630:493–500. 10.1038/s41586-024-07487-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bugnon M, Röhrig UF, Goullieux M et al. (2024) SwissDock 2024: major enhancements for small-molecule docking with Attracting Cavities and AutoDock Vina. Nucleic Acids Res 52:W324–W332. 10.1093/nar/gkae300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grosdidier A, Zoete V, Michielin O (2011) SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res 39:W270–7. 10.1093/nar/gkr366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zoete V, Daina A, Bovigny C et al. (2016) SwissSimilarity: A Web Tool for Low to Ultra High Throughput Ligand-Based Virtual Screening. J Chem Inf Model 56:1399–1404. 10.1021/acs.jcim.6b00174 [DOI] [PubMed] [Google Scholar]
- 44.Meng EC, Goddard TD, Pettersen EF et al. (2023) UCSF ChimeraX: Tools for structure building and analysis. Protein Sci 32:e4792. 10.1002/pro.4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honorato RV, Koukos PI, Jiménez-García B et al. (2021) Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front Mol Biosci 8:729513. 10.3389/fmolb.2021.729513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vangone A, Schaarschmidt J, Koukos P et al. (2019) Large-scale prediction of binding affinity in protein-small ligand complexes: the PRODIGY-LIG web server. Bioinformatics 35:1585–1587. 10.1093/bioinformatics/bty816 [DOI] [PubMed] [Google Scholar]
- 47.Hiller K, Hangebrauk J, Jäger C et al. (2009) MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Analytical chemistry 81:3429–3439. 10.1021/ac802689c [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that data supporting the findings of this study are available within the paper and its supplementary information files. Data derived from single cell sequencing experiments will be made publicly available upon manuscript acceptance.