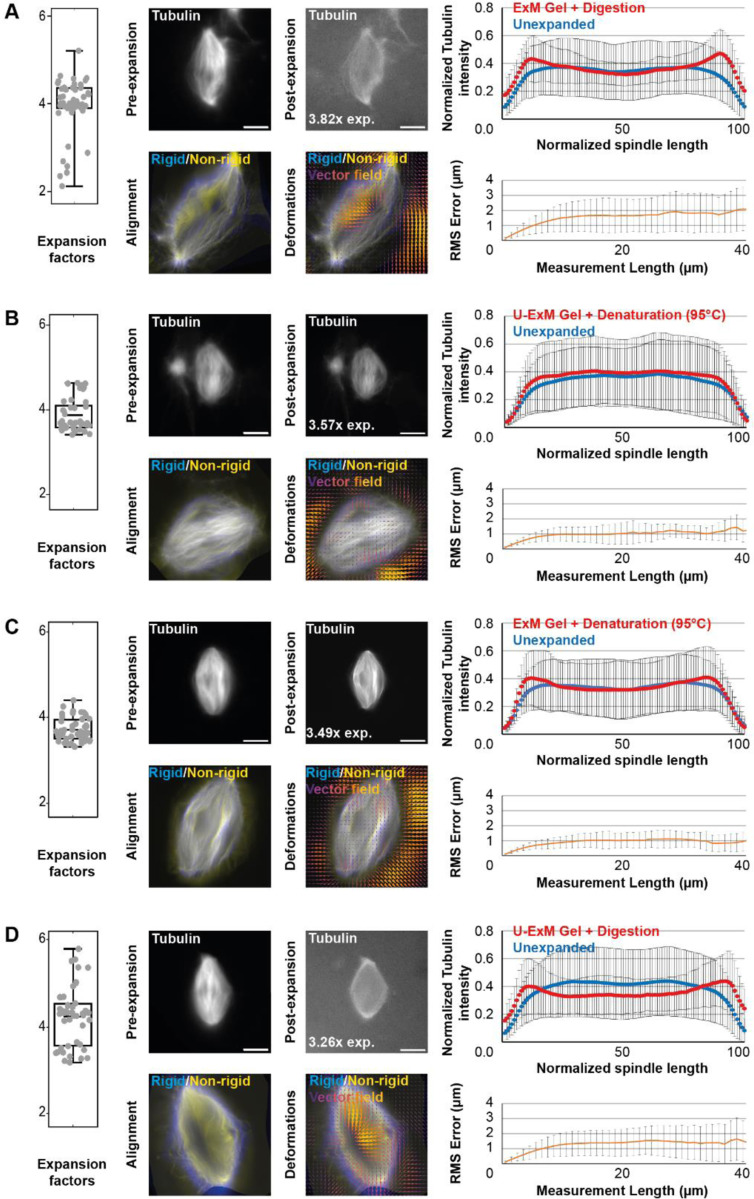
FIGURE 2:
Comparison of expansion factors, rhodamine-labeled tubulin intensity distribution and RMS error between the different expansion protocols. The four tested conditions differing in the acrylamide composition of the gels and the nature of the disruption method are shown: ExM gel + proteinase K digestion (A), U-ExM gel + thermal denaturation (B), ExM gel + thermal denaturation (C), and U-ExM gel + proteinase K digestion (D). Left panels are boxplots showing individual spindle expansion factors (n = 51, n = 41, n = 47, n = 47 spindles from 4 egg extracts respectively for A, B, C, and D), the center line is the mean, the box indicates the 25%−75% quantiles and the error bars the minimum and maximum values. Average expansion factors were A: 3.94 ± 0.63, B: 3.86 ± 0.37, C: 3.71 ± 0.27, D: 4.22 ± 0.67 (average value ± standard deviation). For each, top images are representative images of unexpanded (left) and expanded (right) spindles, with rhodamine-labeled tubulin shown in gray. The expansion factor of the shown spindle is indicated. On the right of the top images are the average normalized fluorescence intensity distributions ± standard deviation for unexpanded (blue) and expanded (red) spindles. Bottom images show the alignment (left) made between rigid registration and non-rigid registration, and (right) the vector field of deformations calculated from the registrations. On the right of the bottom images, panels represent the Root-Mean-Square length measurement error as a function of measurement length for pre- vs. post-expansion images. The orange line shows the average and the error bars, the standard deviation (n = 51, n = 41, n = 47, n = 47 spindles from 4 egg extracts, respectively for A, B, C, and D). Scale bars, 10 μm for unexpanded spindles and 40 μm for expanded spindles.