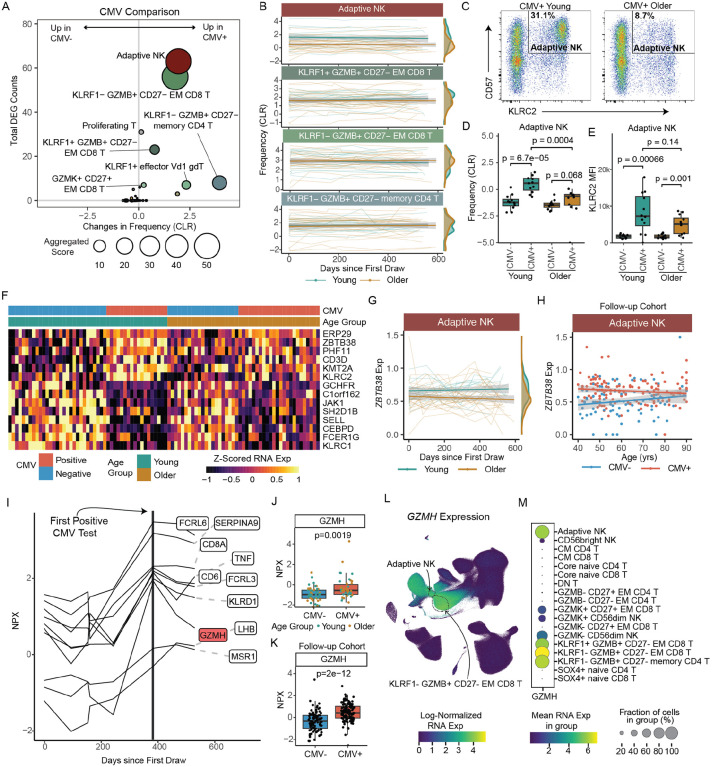
Figure 4. Distinct impact of CMV infection and age on the immune landscape.
A. Bubble plot comparison of the change in frequency (using centered log-ratio (CLR) transformation) and number of DEGs at ‘Flu Vax year 1 day 0’ between CMV+ (n=44) and CMV− (n=52) adults. Bubble size shows a combined metric of change defined as −log10(p.adj from CLR freq comparison) x DEG_Counts. B. Select subset frequencies in PBMCs shown over time. Teal dots are young adults. Bronze dots are older adults. Regression line shown. C. Representative flow plots of CD57 and NKG2C (KLRC2 gene) expression within NK cells. Adaptive NKs cells are defined as CD57+NKG2C+ NK cells. D. Adaptive NK cell frequencies and E. NKG2C (KLRC2) MFI expression on adaptive NKs comparing young CMV− (n=12), young CMV+ (n=12), older CMV− (n=12) and older CMV+ (n=12) adults from spectral flow cytometry analysis. P-values calculated using unpaired Wilcoxon test. F. Heatmap of mean RNA expression levels from upregulated DEGs in adaptive NKs of CMV+ young adults across all individuals in longitudinal cohort (n=96). G. ZBTB38 expression in adaptive NKs (left panel) CMV+ young (n=18) and older (n=24) adults, shown over time (up to 600 days after first blood draw). Teal dots are CMV+ young adults. Bronze dots are CMV+ older adults. Regression line shown. H. ZBTB38 expression in adaptive NKs (right panel) CMV+ (red, n=136) and CMV− (blue, n=98) adults across age in our follow-up cohort. I. Normalized expression (NPX-bridged) of plasma proteins in one young adult who converted from CMV− to CMV+ over the course of our study. Proteins were considered significant if they had a 1.5 or greater fold change pre- to post-conversion. J. Normalized expression of GZMH protein in plasma of CMV− and CMV+ individuals from our longitudinal cohort. Young and older adults are delineated by circle and squares, respectively. K. Normalized expression of GZMH protein in serum of CMV− and CMV+ individuals from our follow-up cohort. P-value for J and K were determined by Wilcoxon rank-sum test. L. UMAP of GZMH RNA expression in PBMCs from all individuals in our longitudinal cohort. M. Dot plot of GZMH RNA expression in NK, CD4 T cell and CD8 T cell subsets.