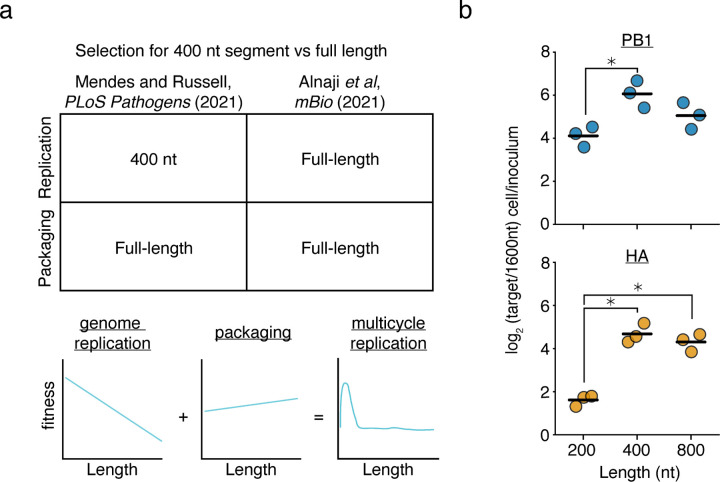
Figure 1. Prior models incompletely describe genome replication during viral infection.
(a) (top)Our previous work found that a 400nt variants of the PB1 and HA segments outcompete their full-length counterparts during genome replication, but during packaging, full-length variants have an advantage. More comprehensive library-based analyses of genomic length showed that a smaller sizes correlate with better replication efficiency and longer sizes better packaging efficiency. Inconsistent with this model, Alnaji et al. described a phenomenon where a short variant of the PB2 segment was outcompeted during genome replication. (bottom) Schematic of our model derived from measuring thousands of length-variants in Mendes and Russell (2021).36 (b) Barcoded 200, 400, 800, and 1600nt variants of A/WSN/1933 PB1 and HA bearing equal sequence length from 5’ and 3’ ends were generated and rescued by coinfection with wild-type virus. Viral supernatant was used to infect A549 cells at an MOI of 25, and qPCR was used to analyze the proportion of each variant before infection and within infected cells at 8h post-infection. 200, 400, and 800nt variants are assessed by their frequency relative to the 1600nt species. Asterisks indicate significantly different values, ANOVA p<0.05 with post-hoc Tukey test, q<0.05. n=3, individual replicates and mean shown.