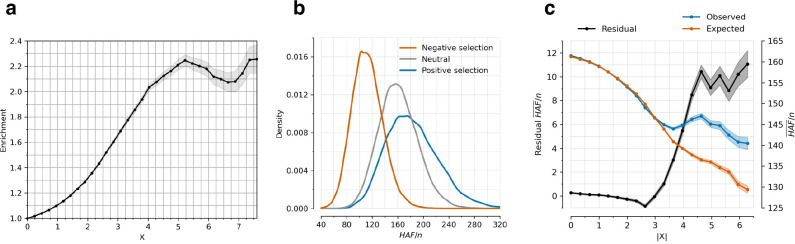
Extended Data Figure 3: Robustness of directional selection signals (related to Figure 1a,b).
(a) Proportion of SNPs significant in any of 452 pan-UK Biobank GWAS studies for X-statistics with magnitudes larger than the threshold on the x-axis, adjusted for minor allele frequency and measures of linked purifying selection (McVicker-B, Murphy-phastCons, and Murphy-CADD). Background selection tends to be higher in functional genomic regions, so SNPs with higher are more penalized than in Figure 1a hence the lower plateau. (b) Simulating neutral, negative, and positive selection for a 200 kb window around a focal SNP, with derived allele frequency drawn uniformly from [0,1]. The focal SNP has , population size is constant at 20000 diploid individuals, mutation rate per base pair per generation is 2×10−8, and recombination rate is 1 cM per 1 Mb. (c) Residual mean (HAF)/n for a haploid sample size n over 200 bp windows is observed minus expected value. Expected value is determined using a linear regression model with McVicker-B, Murphy-phastCons, and Murphy-CADD as variables, providing the expected mean (HAF)/n conditioned on them.