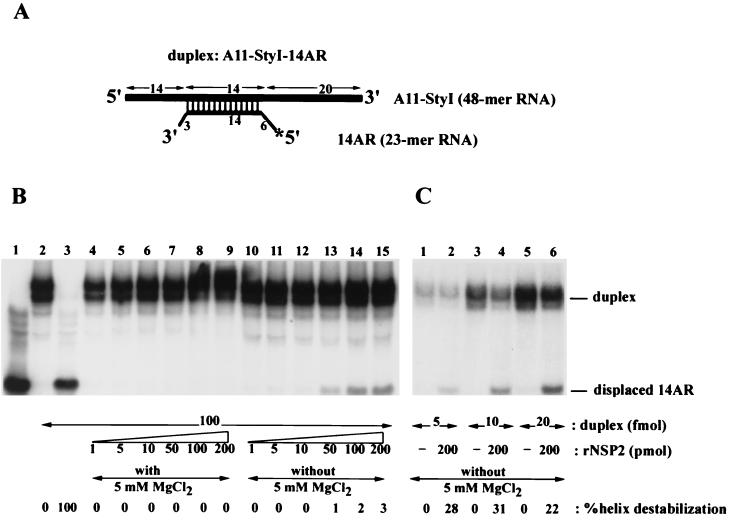
FIG. 8.
Destabilization of an RNA-RNA duplex by NPS2. (A) A schematic representation of the 32P-labeled RNA-RNA duplex A11-StyI–14AR produced by annealing the RNAs A11-StyI and 14AR. (B) Strand displacement assays were performed by incubating 0.1 pmol of the RNA-RNA duplex with 1 to 200 pmol of NSP2 in the presence (lanes 4 to 9) or absence (lanes 10 to 15) of 5 mM MgCl2. As controls, reaction mixtures were also prepared that contained 0.1 pmol of the 14AR RNA instead of the duplex (lane 1), 0.1 pmol of A11-StyI–14AR duplex and no NSP2 (lane 2), or 0.1 pmol of A11-StyI–14AR and no NSP2, denatured by heating at 95°C for 2 min (lane 3). (C) Strand displacement assays were performed by incubating 5 to 20 fmol of the RNA-RNA duplex in the presence or absence of recombinant NSP2 (rNSP2). The reaction mixtures were analyzed by nondenaturing gel electrophoresis and autoradiography. A PhosphorImager was used to determine the percent helix-destabilizing activity for each reaction. The values were normalized to that of 100% for the assay reaction wherein the substrate duplex was denatured by heating.