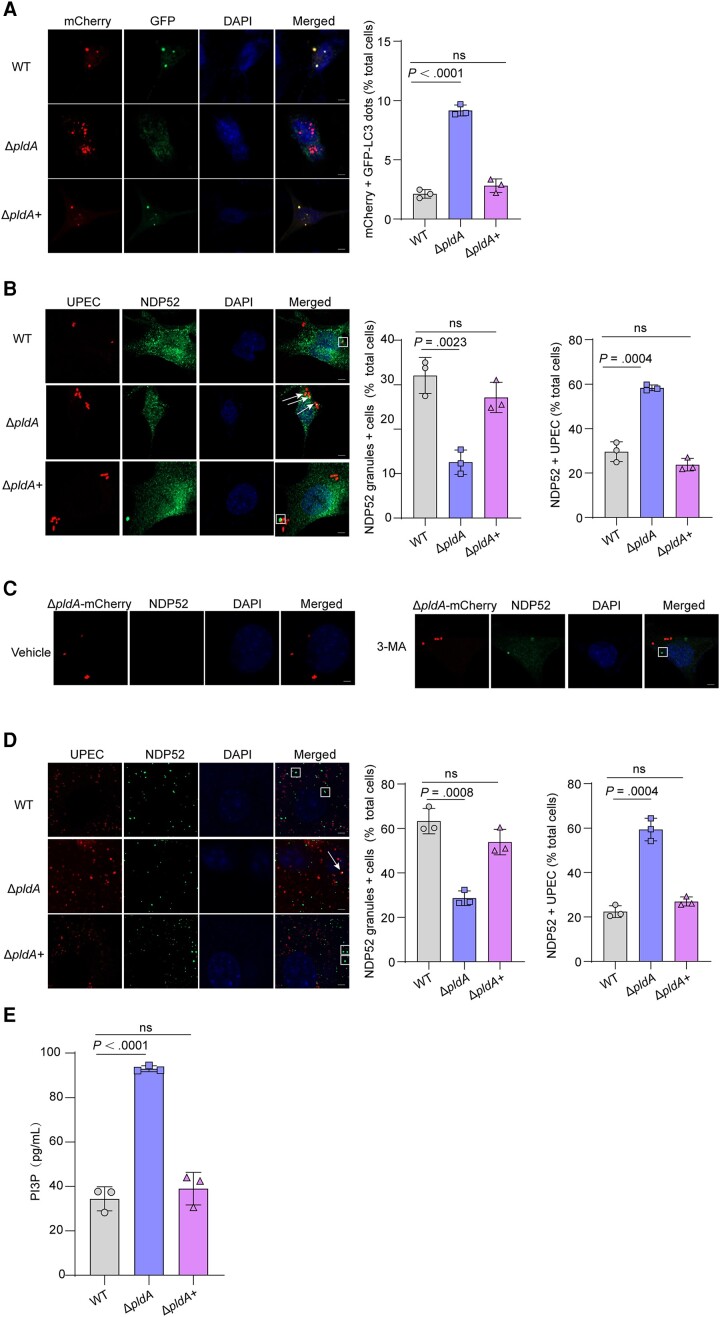
Figure 4.
PldA induces the accumulation of NDP52 granules. A, Representative images of 5637 cells transfected with mCherry-GFP-LC3 for 24 hours and infected with WT, ΔpldA or ΔpldA+ for 4 hours. Autophagosomes, yellow puncta; autolysosomes, red puncta. The bar graph shows the quantification of LC3 red puncta per cell. Scale bar, 5 μm. n = 3 slides. B, Colocalization of NDP52 with CFT073 expressing mCherry in 5637 cells at 4 hpi. The arrows indicate NDP52+ bacteria, and the square frames indicates NDP52 granules (left). The bar graph shows the percentage of cells infected with the indicated strains displaying 1 or several NDP52 granules (middle) and the intracellular bacteria that colocalized with NDP52 relative to the total number of cells (right). Scale bar, 5 μm. n = 3 slides. C, NDP52 granules of ΔpldA-mCherry–infected 5637 cells treated with either vehicle or 2.5 mM 3-MA. The square frame indicates an NDP52 granule. Scale bar, 5 μm. n = 3 slides. D, Colocalization of NDP52 with CFT073 expressing mCherry in mouse bladders at 6 hpi. The arrow indicates NDP52+ bacteria, and the square frame indicates NDP52 granules (left). The bar graph shows the percentage of cells infected with the indicated strains displaying 1 or several NDP52 granules (middle) and the intracellular bacteria that colocalized with NDP52 relative to the total number of cells (right). Scale bar, 5 μm. n = 3 slides. E, 5637 cells infected with WT, ΔpldA, or ΔpldA+ for 4 hours were lysed and PI3P levels were measured by competitive ELISA. Data are presented as the mean ± SD. P values were determined using Student t test. Significance is indicated by the P value. Abbreviations: 3-MA, 3-methyladenine; DAPI, 4′,6-diamidino-2-phenylindole; ELISA, enzyme-linked immunosorbent assay; hpi, hours postinfection; ns, not significant; PI3P, phosphatidylinositol 3-phosphate; RFP-GFP, red fluorescent protein-green fluorescent protein; WT, wild type.