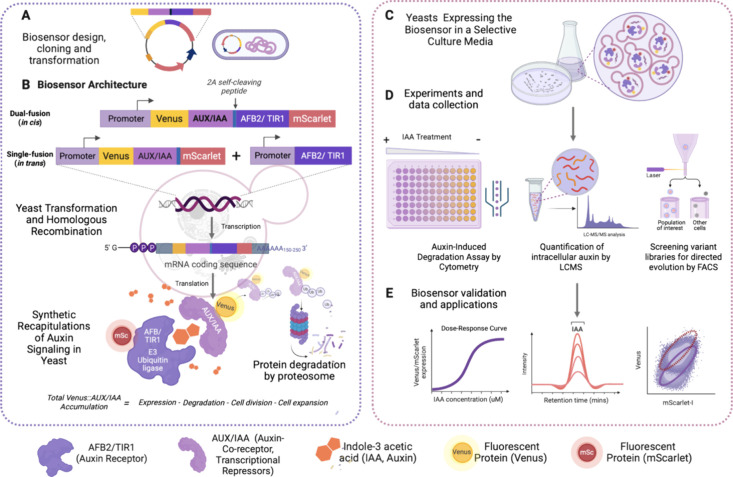
Figure 2.
Schematic illustrating the workflow for engineering genetically encoded auxin biosensors. (A) A plasmid construct for the auxin biosensor is generated, chemically transformed, and amplified in Escherichia coli, and transformed and integrated into the yeast genome by homologous recombination. (B) The dual-fusion (in cis) ratiometric biosensor construct consists of an auxin receptor unit, TIR1 or AFB2, and a coreceptor Aux/IAA, each fused to a fluorescent protein and separated by a 2A self-cleaving peptide. Single-fusion (in trans) constructs have TIR1 or AFB2 expressed in trans to the ratiometric fluorescent reporter. Expression of these biosensors generates synthetic recapitulations of plant auxin signaling and auxin-induced Aux/IAA fusion protein degradation. The biosensor response is measured by the ratio of TIR1/AFB2-mScarlet-I (or free mScarlet-I) to Venus-Aux/IAA, which is proportional to the auxin concentration at a given time point. (C, D) Positive biosensor-expressing yeast colonies were inoculated into synthetic growth media and incubated overnight for auxin-induced degradation assays via flow cytometry. (E) The capability of the biosensor to detect and quantify auxin was analyzed by comparing the biosensor response (ratio of Aux/IAA-fused to free or TIR1/AFB2-fused fluorescent proteins) to intracellular auxin measurements via LC-MS. The biosensor may also be used to measure functional variation of mutants in TIR1/AFB or Aux/IAA genes with greater precision than prior methods.