Abstract
Milk samples from 50 sheep on a single Scottish research farm were collected weekly for 10 wk postpartum. Samples were analyzed for somatic cell counts (SCC) each week and bacteriologic culture was done for 7 of the 10 wk. A total of 492 udder half samples were cultured, of which 467 had corresponding cell count data. Statistical analysis on complete SCC and culture data showed no association between SCC and bacterial isolation, even when more than 10 colonies of a single bacterial species were present. Only 3.6% of the samples were simultaneously positive for high count (> 10 colonies from 0.01 mL of milk) of any one bacterial species and high SCC (> 1 × 106/mL). The bacteria recovered were: Staphylococcus equorum (19 times), S. xylosus (7 times), S. simulans (6 times), Streptococcus uberis (3 times) and other streptococci (4 times), Mannheimia (Pasteurella) haemolytica (2 times), Staphylococcus aureus (1 time), S. capitis (1 time), and Enterococcus faecium (1 time). There was an association between the test day and SCC, with higher SCC values in the first 2 wk. In addition, significantly higher SCC values were found in the oldest animals compared to the other age groups.
Résumé
Des échantillons de lait provenant de 50 brebis d’une ferme expérimentale écossaise ont été prélevés à chaque semaine pendant 10 semaines post-partum. Le comptage des cellules somatiques (SCC) a été fait sur des échantillons prélevés à toutes les semaines alors que la culture bactérienne a été effectuée sur des échantillons de 7 des 10 semaines. Sur un total de 492 échantillons mis en culture 467 avaient des données de SCC correspondantes. Les analyses statistiques des SCC et des résultats de culture ne montraient pas d’association entre l’isolement bactérien et le SCC, et ce même si plus de 10 colonies d’une même espèce étaient présentes. Seulement 3,6 % des échantillons étaient positives simultanément pour un dénombrement élevé d’une même espèce bactérienne (> 10 colonies/0,01 mL de lait) et un SCC élevé (> 1 × 106/mL). Les bactéries retrouvées étaient : Staphylococcus equorum (19 fois), S. xylosus (7), S. simulans (6), Streptococcus uberis (3) et autres streptocoques (4), Mannheimia (Pasteurella) haemolytica (2), S. aureus (1), S. capitis (1) et Enterococcus faecium (1). Une association a été notée entre le jour du test et le SCC, avec des valeurs de SCC plus élevées durant les 2 premières semaines. De plus, des valeurs de SCC significativement plus élevées ont été trouvées chez les animaux plus âgés comparativement aux autres groupes d’âge.
(Traduit par Docteur Serge Messier)
Introduction
Microbial analysis and somatic cell counts (SCC) have been used to diagnose subclinical mastitis (SCM) in ewes. This disease has received little attention, particularly in places where sheep are raised for meat rather than milk. Subclinical mastitis is thought to have a prevalence of between 10% and 30% in lowland flocks in Southern England (1,2). This condition can affect milk yield and milk composition, with consequent effects on growth rates of lambs (3,4). Comparative studies on ovine SCM are hampered because of the lack of universally accepted standards for the diagnosis of this problem. Colony counts of various bacterial isolates in combination with a high SCC or bacterial colony count, or SCC alone, are used for case definition (5). Fthenakis (6) used the term SCM for cases where 0.01 mL of a milk sample yielded > 10 colonies of a single organism on Columbia Blood Agar, and simultaneously the sample had a SCC of > 1.0 × 106 cells/mL. Other workers (7,8) have used lower threshold values for the diagnosis of SCM. In bovine mastitis, major pathogens are considered of relevance when fewer colonies are found. The National Mastitis Council (NMC) guidelines consider a single colony of Staphylococcus aureus or Streptococcus agalactiae significant in culture. Environmental streptococci must have 2 to 9 colonies to be considered significant, whereas, coagulase-negative staphylococci (CNS) must have > 10 colonies in pure culture to be considered relevant (9).
The objectives of this study were to gain information on levels of SCM in a population of ewes by SCC and bacteriological analysis of milk obtained by repeated sampling during a period of 10 wk postlambing, and to examine the association between culture status and SCC.
Materials and methods
Fifty Scottish Blackface cross Border Leicester (Greyface) ewes of 2 to 7 y of age from Moredun Research Institute’s farm located in Midlothian, Scotland, were used in this study. The group was randomly selected from a 500 ewe breeding flock set up to provide animals for use in Institute studies, as well as for commercial lamb production. To allow easy gathering for weekly milk sampling, the sheep used in the study were maintained as a single group. All ewes were housed in individual pens for approximately 24 h after lambing followed by housing in open pens for 3 to 7 d before being turned out to graze with their lambs for the remainder of the experiment. Ewes with clinical mastitis were not included in this study. In compliance with the United Kingdom (UK) Animals (scientific procedures) Act of 1986, all required approvals were obtained prior to the experiments. From 3 d postlambing, 10 to 15 mL milk samples were obtained from each udder at weekly intervals for 10 wk. The sheep were restrained in a sitting position in a cradle that exposed the udder. The foremilk was removed and the teats were then wiped with disposable alcohol impregnated swabs (Sagrosept wipes; Schulke and Mayr, Sheffield, UK) before a mid-flow milk sample was manually expressed from each udder into separate sterile 25 mL universal tubes (Bibby Sterilin, Staffs, UK). The samples were examined immediately or held at 4°C for less than 24 h. Samples for all 10 wk were examined for SCC using a Coulter counter (Coulter Counter ZM; Beckman Coulter, Buckinghamshire, UK). The machine was calibrated for bovine SCCs, rechecked prior to each sample run, and the samples processed in accordance with the methods described by the International Dairy Federation (10). Briefly, the 10 mL milk samples were fixed in 3.5% formaldehyde and diluted in 1:400 in a buffered isotonic saline solution (Lechesol; Apertech, Bedfordshire, UK), and a 0.5 mL aliquot was subsequently used for SCC measurement. To correct for sample dilution, the number of SCC/mL was obtained by multiplying the SCC data by 800 ([(Raw SCC × 400)/500] × 1000).
Samples obtained in the 1st, 2nd, 5th, 6th, 8th, 9th, and 10th wk were also examined for presence of bacteria by culture. A 0.01 mL sample of milk was plated on Columbia blood agar and MacConkey agar number 2 (Oxoid, Basingstoke, UK), using a calibrated loop. The plates were incubated aerobically at 37°C for up to 72 h, examined daily for the number and types of colonies, and the information recorded. Bacteriological interpretation was based on the NMC recommendation for bovine milk cultures (9). Briefly, samples were considered to have a major pathogen if they had 1 or more colonies of Staphylococcus aureus, Streptococcus agalactiae, or Mannheimia (Pasteurella) haemolytica; if they had 2 or more colonies of environmental streptococci growing in pure culture; or more than 10 colonies in mixed culture with one other species. Samples were considered to have a minor pathogen if greater than 10 colonies of CNS, corynebacteria, or other bacteria, excluding Bacillus species, were present. Samples with 3 or more species on a plate were considered contaminated. The bacteria were identified based on colony morphology; Gram’s stain; and tests as required, for catalase, coagulase, oxidase, and finally, using API bacterial identification strips (“ID32 STAPH,” “rapid ID32 STREP,” “api Coryne,” “api 20 NE;” bioMerieux sa, Lyon, France).
A total of 233 samples comprising all of the 2nd and 3rd wk (total 187), and 46 samples from the 10th wk (selected on the basis of SCC > 1 × 106 in 1 or more previous weeks) were, in addition, selectively cultured for Listeria monocytogenes as per the methods described by Fthenakis and others (11). In brief, milk samples were cultured in Listeria broth with selective supplement (Oxoid CM 862, SR 141; Oxoid) and, after incubation at 30°C, subcultured onto supplemented polymyxin B-acriflavine-lithium chloride-ceftazidimeaesculin-mannitol (PALCAM) agar (Oxoid), and examined for Listeria colonies.
All data were transferred to a spreadsheet program (Quattro Pro, version 8.0; Corel Corporation, Ottawa, Ontario) and exported to a statistical package (Stata, version 7.0; Stata Corporation, College Station, Texas, USA) for analysis. The data entered into the program included the identification number of the sheep, year code, origin of the sample (left or right udder), laboratory identification number, sample date, SCC, bacterial growth, number of colonies, and bacterial identification.
Descriptive statistics for bacterial growth and SCC parameters were created. For analysis, SCC data was converted to the natural log (LNSCC) of the raw SCC value. Linear regression models were constructed to examine associations between the LNSCC and bacteria classified into 2 groups for the purpose of major and minor pathogens, as previously described. Analysis was done at 3 cutoff points of SCC (500 000, 1 million, and 2 million). Additional regression models examined the association between test day, udder half, birth year (ear tag code), and LNSCC.
Results
A total of 492 milk samples were cultured during the study period, of which 467 had corresponding cell count data. The results of culture are summarized in Table I. Of these, 178 (38.1%) were negative for bacterial growth and 117 (25.1%) were contaminated. Ten samples were positive for major pathogens, which included 1 Staphylococcus aureus, 3 Streptococcus uberis, 4 Streptococcus spp., and 2 Mannheimia (Pasteurella) haemolytica. Thirty-six were positive for minor pathogens, which included 19 Staphylococcus equorum, 7 S. xylosus, 6 S. simulans, 1 S. capitis, 1 Enterococcus faecium, 1 Corynebacterium spp., and 1 Aerococcus spp. Ninety-four samples had mixed growth with < 10 colonies of minor pathogen types, and 20 samples had < 10 colonies of CNS. Twelve others had growth of Bacillus (8 times), alpha-hemolytic streptococci (1), Enterococcus spp (1), and unidentified Gram-negatives (2). None of the 233 samples examined was positive for growth of Listeria monocytogenes.
Table I.
Culture results of 467 milk samples with high somatic cell counts (SCC)a
| Samples with
|
||||
|---|---|---|---|---|
| Growth/Bacterium | Number | % | High SCC | % |
| No growth | 178 | 38.1 | 70 | 39.3 |
| Contaminated with Bacillus | 117 | 25.1 | 55 | 47.0 |
| Major pathogen | 10 | 2.1 | 2 | 20.0 |
| Minor pathogen | 36 | 7.7 | 15 | 41.6 |
| Mixed | 94 | 20.1 | 54 | 57.4 |
| CNS < 10 colonies | 20 | 4.3 | 13 | 65.0 |
| Other | 12 | 2.5 | 7 | 58.3 |
CNS — Coagulase-negative staphylococci
SCC/mL = > 1 × 106 for the sample
The SCC value was < 1 × 106/mL for most of the culture-positive samples, including 1 positive for Staphylococcus aureus and 1 positive for M. haemolytica. However, the mean SCC values for 10 wk, from the samples originating from the S. aureus and M. haemolytica-positive udder halves, were > 1 × 106 (data not presented). Overall, only 17 (3.6%) of 467 samples were simultaneously positive for bacteriologic culture (major and minor pathogens) and SCC (> 1 × 106 cell/mL).
Nineteen samples were positive (> 10 colonies) for Staphylococcus equorum. Of these, only 6 samples were positive by SCC, whereas 5 of the 7 S. xylosus-positive samples had high SCC values.
Regression analysis, using the 467 culture results with SCC data, did not reveal any association between the LNSCC and pathogens, either those considered major (n = 10) or minor (n = 36) (4 of the minor pathogens did not have SCC’s). The SCC data are summarized in Table II. Figure 1 illustrates the LNSCC and the week for culture negative versus positive (major or minor pathogens). The mean SCC for all samples of the study, regardless of whether there was a corresponding bacterial culture was 1867 × 103. The mean SCC of samples that were bacterial culture-positive was 2351 × 103. The mean SCC of samples that were negative for bacterial growth was 2285 × 103. No association between bacterial culture positivity and SCC was found. The mean SCC value for major pathogens was 1762 × 103, and for the 36 samples positive for minor pathogens, it was 2293 × 103. There was no statistically significant difference (confidence level 0.95) between these values and the value for samples with no bacterial growth. There was an association between test day and LNSCC. Samples in the first 2 test weeks (mean = 3426.22 × 103) had higher SCC (P < 0.01) than those on the other test days (mean = 971.74 × 103). There was no association between SCC and udder half. No association was found between the SCC of noninfected glands and the infection status of the contra-lateral gland. When ear tag code (birth year) was examined, the oldest animals (birth year X) had higher SCC than those in other age groups (P < 0.05).
Table II.
Summary of somatic cell count (SCC) data ×103
| Cell countsa | All weeks culture | Weeks with negative | Culture pathogen | Major pathogen | Minor pathogen |
|---|---|---|---|---|---|
| Number of observations | 925 | 467 | 178 | 10 | 36 |
| Mean | 1876 (6.98)b | 2351 (7.12) | 2285 (7.03) | 1762 (6.77) | 2293 (7.17) |
| Median | 933 (6.84) | 944 (6.85) | 905 (6.81) | 673 (6.51) | 902 (6.80) |
| Standard deviation | 4053 (0.84) | 4907 (0.92) | 5366 (0.92) | 3371 (0.97) | 3067 (1.01) |
| Minimum | 113 (4.73) | 174 (5.16) | 213 (5.36) | 371 (5.92) | 256 (5.45) |
| Maximum | 50000 (10.82) | 50000 (10.82) | 47286 (10.77) | 11329 (9.34) | 14702 (9.60) |
SCC/mL
Linear scores
Figure 1.
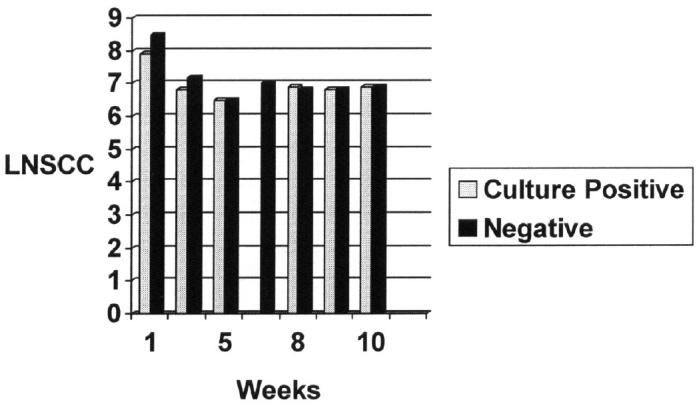
Natural log of raw somatic cell count (LNSCC) values versus culture results.
Milk samples from the left udder half of 2 ewes were repeatedly (3×) positive for 2 species of bacteria (Table III). Streptococcus uberis was isolated (> 50 colonies) for 3 wk from samples obtained from the left udder half of ewe 1655Z-L, but the SCC was positive only in the 1st wk. Similarly, a second ewe was positive for Staphylococcus equorum, with positive SCC only in the 1st wk. However, the mean SCC values for 10 wk in both cases were > 1 × 106/mL.
Table III.
Somatic cell counts (SCC) compared to repeated bacterial isolation Somatic cell
| Somatic cell counts × 103 for the 3 weeks
|
|||||
|---|---|---|---|---|---|
| Sheep per sample | Pathogen | 1 | 2 | 3 | Mean weeks 1 to 10 |
| 1655Z-L | Streptococcus uberisa | 9063 | 467 | 444 | 1421 |
| (9.11 | 6.15 | 6.10)c | (6.99) | ||
| 1 | 5 | 9 | |||
| 1317B-L | Staphylococcus equorumb | 8186 | 564 | 480 | 1648 |
| (9.01 | 6.34 | 6.17) | (6.92) | ||
> 100 colonies week 1, > 50 colonies week 2, > 200 colonies week 9
10 to 30 colonies all 3 weeks
Linear scores in brackets
Discussion
The diagnosis of SCM on the basis of a selected SCC value or a particular colony count of bacteria is problematic because workers have used different cut-off values. There is a lack of an accepted “normal” value, and there is no agreement on the accepted number of colonies as significant or vice versa. A high SCC value in the absence of bacterial growth on blood agar may be due to bacteria, such as mycoplasma, or due non-bacterial causes, including physiological factors. Perinatal SCC values were higher, which could be explained by these physiologic factors. It is also possible that SCC values may change with the stage of bacterial infection. In the present study, milk from the same sheep was sampled for 10 wk for SCC, and bacterial culture examined 7 times during the 10-week period. Statistical analysis on complete SCC and bacterial culture data showed no association between SCC and bacterial isolation, even when more than 10 colonies of a fully identified single bacterial species were present. However, 3.6% of 467 samples were simultaneously positive for a bacterial pathogen and SCC. Using the definition of SCM as the presence of both bacteriologically positive and SCC positive results, Jones and Watkins (2) reported a 11.7% rate of SCM during lactation among the lowland flocks in England and Wales. The 3.6% prevalence rate of SCM among the 50 ewes at the Firth Mains farm in this study is quite low compared to the study mentioned above. To determine the infection rate in a herd, both bacteriological status and SCC in milk should be taken into account, especially when prevalence within a herd is not known (12).
Thirty-three of 492 (6.7%) samples were positive for CNS and S. equorum, followed by S. xylosus and S. simulans. Coagulasenegative staphylococci are the most frequently isolated bacterial group from ewes with SCM and during lactation (4–7). Although S. equorum isolates in the present study had excellent identification under the API Staph Ident system, reports on this species are scanty and it has only been isolated from goat milk (13). Staphylococcus epidermidis, S. simulans, S. sciuri, and S. xylosus were the most common isolates from SCM in ewes in Greece (6). Burriel (14) reported that S. simulans, S. xylosus, and S. hyicus were the predominant species of CNS in the milk from meat ewes, and S. epidermidis was the predominant species in the milk of dairy ewes. However, in the present study, the API Staph system, which is recommended by the National Mastitis Council (15) for species-level identification of CNS, was used. It should be noted that some variations in speciation of CNS can occur when different commercial identification systems are used (16).
The majority of our isolates were CNS. In a study conducted by Gonzalez-Rodriguez (7) CNS were the most frequently isolated bacterial group, although these bacteria gave rise to significantly lower SCC values compared with coagulase-positive staphylococci and streptococci. Fthenakis and Jones (3) induced SCM in ewes with a strain of Staphylococcus simulans, and noted an increase in SCC following infection. In our study, 6 samples were positive (> 10 colonies) for S. simulans. No association was found between culture positivity for this bacterium and increased SCC, though in 1 sample the SCC was high. Similarly, some of the samples positive for Staphylococcus equorum and S. xylosus had high SCC values, but a statistical association was not found between culture positivity and SCC. It is possible that species of CNS and strains under each species have varying pathogenic properties. Some reasons for a lack of correlation between bacterial culture positivity and SCC have been pointed out and include the coexistence of bacteria in the absence of inflammation, lack of actual parenchymal infection, and use of different equipment for performing SCC (2,6,17).
Only 1 of the 2 samples that were positive for M. (P.) haemolytica had a high SCC. Mannheimia (Pasteurella) haemolytica strains from different sources, including isolates from cases of SCM, have been shown to vary in virulence, and in their ability to establish colonies in the mammary gland (18).
The SCC data in Table III seems to question any stable relationship between presence of bacteria in large numbers and SCC. Despite high counts of Streptococcus uberis, in samples obtained within 3 wk, there was no high SCC except for the 1st wk. Further, there is strong evidence of intermittent excretion of the bacterium because of the fact that the samples from only weeks 1, 2, and 9 were positive (50 to 200 colonies), while samples obtained in the other weeks were bacteriologically negative. This could also be interpreted to be selfcure and reinfection. Fhenakis (6) has discussed the dynamics of SCM, including intermittent excretion of bacterial pathogens in milk from ewes. Ewes have been shown to develop SCM with Mycoplasma bovigenitalium, characterized by intermittent Mycoplasma excretion and low milk cell levels (19). The absence of increased SCC with the occurrence of heavy bacterial growth with Streptococcus uberis is puzzling and needs further research.
Naturally occurring ovine SCM is a matter of concern and recent reports implicate bacteria other than the conventional pathogens, including Listeria monocytogenes (11) (which was not isolated from any samples in the present study), Corynebacterium mastitidis and C. camporealensis (2 newly reported species) (20,21), Streptococcus parasanguinis (22), and Burkholderia cepacia (23).
Although the occurrence of Bacillus spp. in milk samples from ewes can be high at times, the contamination rate of 25% found in the present study needs further investigation as to the possible role of the environment. Watson and Buswell (24) noted that Bacillus spp. is predominant in the milk samples from ewes on pasture.
In conclusion, this study did not demonstrate any valid association between positivity for bacteria by culture and SCC in the 492 milk samples from ewes studied during a period of 10 wk of lactation. If one considers simultaneously positive results for significant bacterial growth (> 10 colonies of a particular species from 0.01 mL of milk) and high SCC (> 1 × 106/mL), only 3.6% of samples were positive for SCM, a very low prevalence among the ewes in this study. There was an association between the test day and SCC, with higher SCC in the first 2 wk. In addition, significantly higher SCC values were found in the oldest animals compared to the other age groups.
Acknowledgments
The authors thank Roy Davie and Malcolm Quirie of the Moredun Institute for sample collection and general assistance, and Theresa Rogers of the Atlantic Veterinary College for entering culture data into the spread sheet program.
References
- 1.Watkins GH, Burriel AK, Jones JET. A field investigation of subclinical mastitis in sheep in southern England. Br Vet J. 1991;147:413–420. doi: 10.1016/0007-1935(91)90083-Y. [DOI] [PubMed] [Google Scholar]
- 2.Jones JET, Watkins GH. Studies on mastitis in sheep at the Royal Veterinary College. Sheep Vet Soc Proc. 1998;22:83–90. [Google Scholar]
- 3.Fthenakis GC, Jones ET. The effects of experimentally induced subclinical mastitis on milk yield of ewes and on the growth of lambs. Br Vet J. 1990;146:43–49. doi: 10.1016/0007-1935(90)90075-E. [DOI] [PubMed] [Google Scholar]
- 4.Burriel AR. Dynamics of intramammary infection in the sheep caused by coagulase-negative staphylococci and its influence on udder tissue and milk composition. Vet Rec. 1997;40:419–423. doi: 10.1136/vr.140.16.419. [DOI] [PubMed] [Google Scholar]
- 5.Kirk JH, Glenn JS. Mastitis in ewes. Compend Contin Educ Pract Vet. 1996;18:582–590. [Google Scholar]
- 6.Fthenakis GC. Prevalence and aetiology of subclinical mastitis in ewes of Southern Greece. Small Rumin Res. 1994;13:293–300. [Google Scholar]
- 7.Gonzalez-Rodriguez MC, Gonzalo C, San-Primitivo F, Carmenes P. Relationship between somatic cell count and intramammary infection of the half udder in dairy ewes. J Dairy Sci. 1995;78:2753–2759. doi: 10.3168/jds.s0022-0302(95)76906-5. [DOI] [PubMed] [Google Scholar]
- 8.Gonzales-Rodriguez MC, Carmenes P, Rodriguez MCG. Evaluation of the California mastitis test as a discriminant method to detect subclinical mastitis in ewes. Small Rumin Res. 1996;21:245–250. [Google Scholar]
- 9.National Mastitis Council Inc. Laboratory and Field Handbook on Bovine Mastitis. WD Hoard & Sons Co, Fort Atkinson, Wisconsin, USA. 1987:151.
- 10.International Dairy Federation. Recommended methods for somatic cell counting in milk. Bull Internat Dairy Fed. 1984;168:1–19. [Google Scholar]
- 11.Fthenakis GC, Saratsis P, Tzora A, Linde, K Naturally occurring subclinical ovine mastitis associated with Listeria monocytogenes. Small Rumin Res. 1998;31:23–27. [Google Scholar]
- 12.McDougall S, Murdough P, Pankey W, Delaney C, Barlow J, Scruton D. Relationship among somatic cell count, California mastitis test, impedance and bacteriological status of milk in goats and sheep in early lactation. Small Rumin Res. 2001;40:245–254. doi: 10.1016/s0921-4488(01)00185-7. [DOI] [PubMed] [Google Scholar]
- 13.Meugnier H, Bes M, Vernozy-Rozand C, et al. Identification and ribotyping of Staphylococcus xylosus and Staphylococcus equorum strains isolated from goat milk and cheese. Internat J Food Microbiol. 1996;31:325–331. doi: 10.1016/0168-1605(96)00975-0. [DOI] [PubMed] [Google Scholar]
- 14.Buriel AR. Isolation of coagulase-negative staphylococci from the milk and environment of sheep. J Dairy Res. 1998;65:139–142. doi: 10.1017/s0022029997002689. [DOI] [PubMed] [Google Scholar]
- 15.National Mastitis Council. Laboratory Handbook on Bovine Mastitis (Revised Edition). National Mastitis Council Inc, Madison, Wisconsin, USA. 1999:81.
- 16.Burriel AR, Scott M. A comparison of methods used in species identification of coagulase-negative staphylococci isolated from milk of sheep. Vet J. 1998;155:183–188. doi: 10.1016/s1090-0233(98)80017-8. [DOI] [PubMed] [Google Scholar]
- 17.Burriel AR. Evidence of breed susceptibility to experimentally produced ovine subclinical mastitis. Sheep Goat Res J. 1997;13:20–23. [Google Scholar]
- 18.Watkins GH, Jones JET. The effect of the intramammary inoculation of lactating ewes with Pasteurella haemolytica isolates from different sources. J Comp Pathol. 1992;106:9–14. doi: 10.1016/0021-9975(92)90063-z. [DOI] [PubMed] [Google Scholar]
- 19.Ball HJ. Experimental mastitis caused by Mycoplasma bovigenitalium and M. canadense in the ewe. Vet Microbiol. 1990;22:383–388. doi: 10.1016/0378-1135(90)90025-q. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Garayzabal JF, Collins MD, Hutson RA, et al. Corynebacterium mastitidis sp. nov., isolated from milk of sheep with subclinical mastitis. Internat J Syst Bacteriol. 1997;47:1082–1085. doi: 10.1099/00207713-47-4-1082. [DOI] [PubMed] [Google Scholar]
- 21.Fernandes-Garayzabal JF, Collins MD, Hutson RA, Gonzalez I, Fernandez E, Doninguez L. Corynebacterium comparalensis sp. nov., associated with subclinical mastitis in sheep. Internat J Syst Bacteriol. 1998;48:463–468. doi: 10.1099/00207713-48-2-463. [DOI] [PubMed] [Google Scholar]
- 22.Fernandes-Garayzabal JF, Fernandez E, Las Heraa A, Pascual C, Collins MD, Doninguez L. Streptococcus parasanguinis: New pathogen associated with asymptomatic mastitis in sheep. Emerg Infect Dis. 1998;4:645–647. doi: 10.3201/eid0404.980417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berriatua E, Ziluaga I, Miguel-Virto C, et al. Outbreak of subclinical mastitis in a flock of dairy sheep associated with Burkholderia cepacia complex infection. J Clin Microbiol. 2001;39:990–994. doi: 10.1128/JCM.39.3.990-994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson DJ, Buswell JF. Beecham mastitis series. Modern aspects of sheep mastitis. Br Vet J. 1984;140:529–534. doi: 10.1016/0007-1935(84)90003-4. [DOI] [PubMed] [Google Scholar]


