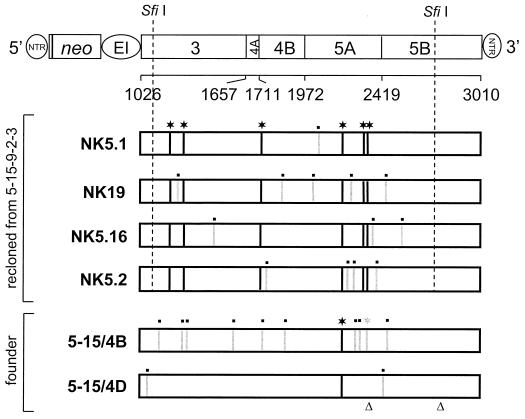
FIG. 1.
Sequence analysis of HCV replicons isolated from various cell lines. The structure of the selectable replicon construct is shown in the top, with numbers below the NS3-NS5B polyprotein referring to the P1 positions of the cleavage sites or its carboxy terminus. neo, neomycin phosphotransferase gene; EI, EMCV IRES. Since for optimal HCV IRES activity ∼36 nt of the core ORF are required, 12 amino acid residues of the core protein are fused to the amino terminus of the neomycin phosphotransferase. Coding regions of the NS3-NS5B polyprotein cloned from cell line 5-15-9-2-3 and the founder line 5-15 are drawn below. Differences in the amino acid sequences are indicated by vertical lines. A black line labeled with a star refers to an amino acid substitution that was conserved with the four sequences isolated from 5-15-9-2-3 cell lines, and gray lines labeled with a dot indicate nonconserved mutations. Note that both replicons cloned from the founder cell line 5-15 carried the proline substitution in NS5A at position 2197 but that the glutamate substitution in NS5A at position 2350 (labeled with a gray star) was found in only one of these replicons (clone 5-15/4B). The positions of single nucleotide deletions in the polyprotein coding sequences of clone 5-15/4D are indicated with triangles.