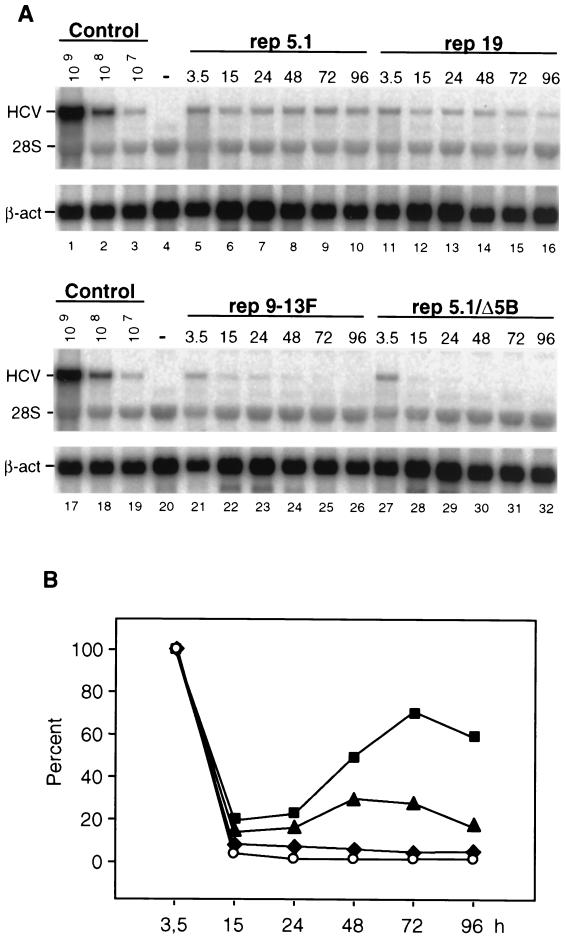
FIG. 3.
Transient replication of cell culture-adapted HCV replicons. (A) Huh-7 cells were transfected with selectable replicon RNAs and harvested at time points indicated above each lane. HCV RNAs were analyzed by Northern blotting. The number of replicon RNA molecules was determined by comparison with the serial dilution of in vitro transcripts (lanes 1 to 3). β-Actin RNA (β-act) served as a control to correct for the amount of total RNA loaded in each lane of the gel (∼2 μg). The result obtained with total RNA from naïve Huh-7 cells is shown in lanes 4 and 20. The positions of replicon RNAs, 28S RNA, and β-actin m-RNA are given in the left. The replicon carrying a deletion that spans the active site of the NS5B RdRp served as a negative control (rep5.1/Δ5B). (B) Quantification of the RNA in the Northern blots shown in panel A by phosphorimaging. Values were corrected for the RNA amounts determined 3.5 h after transfection, and they are expressed as percentages. With rep5.1, the 100% value corresponds to 1.3 × 107 replicon RNA molecules per μg of total RNA. ■, replicon 5.1; ▴, replicon 19; ⧫, replicon 9-13F; ○, replicon 5.1/Δ5B. Analogous results were obtained in three further independent experiments.