Figure 2. Transcriptional diversity of CD8 T cells.
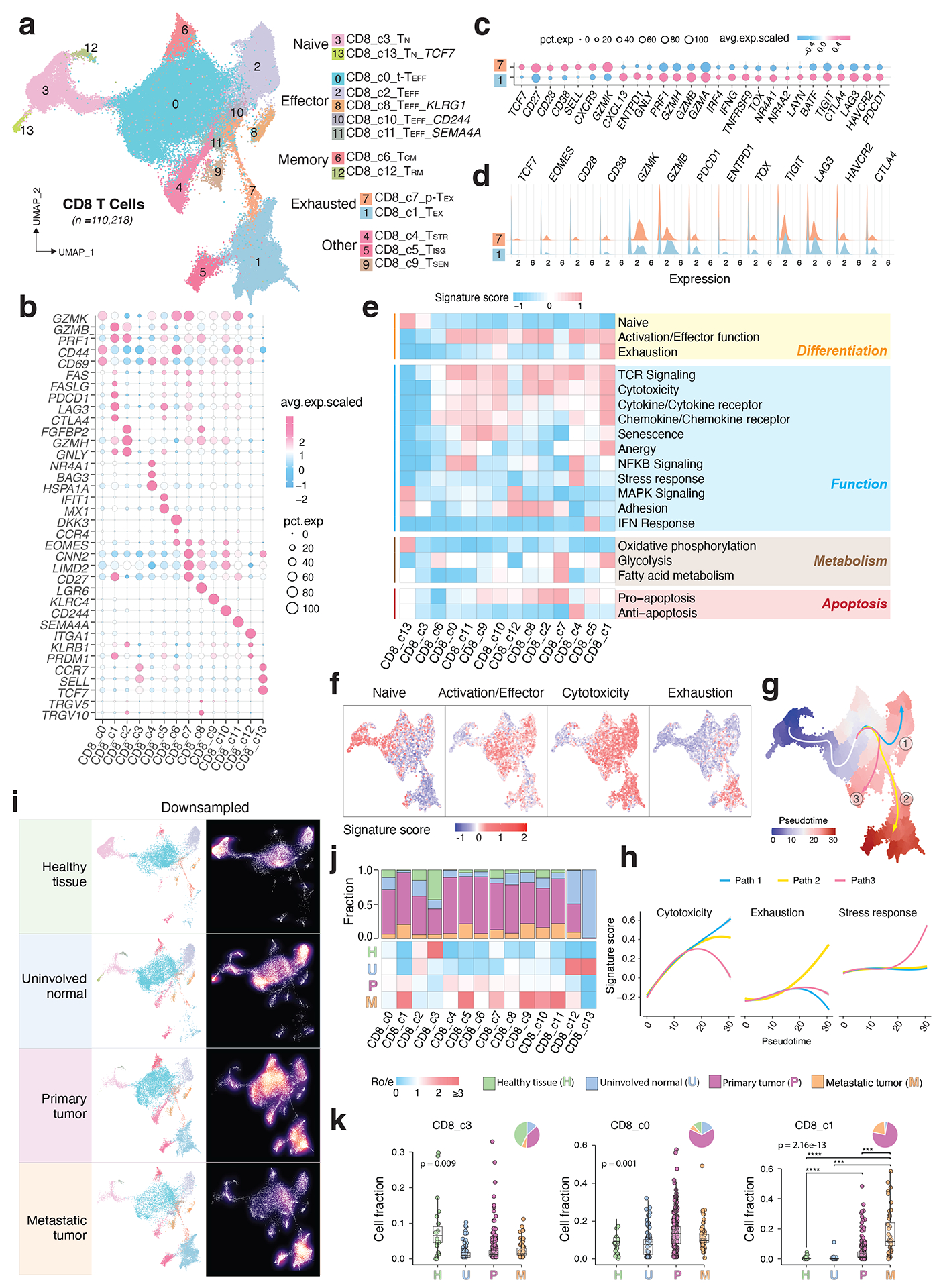
a) The UMAP view of 14 CD8 T cell clusters. b) Marker gene expression across defined T cell clusters. Bubble size is proportional to the percentage of cells expressing a gene and color intensity is proportional to average scaled gene expression. c) Bubble plot and d) ridge plot showing key marker gene expression between the two CD8 T cell clusters. e) Heatmap illustrating expression of 19 curated gene signatures across CD8 T cell clusters. Heatmap was generated based on the scaled gene signature scores. f) Expression of 4 representative gene signatures selected from e). g) Monocle 3 trajectory analysis of CD8 T cell differentiation revealing three main divergent trajectories. Cells are color coded for their corresponding pseudotime. h) Two-dimensional plots showing expression scores for 3 representative gene signatures in cells of paths 1 (blue), path 2 (yellow), and path 3 (pink), respectively, along the inferred pseudotime. i) UMAP view of CD8 T cell states (left) and cell density (right) displaying CD8 T cell distribution across 4 main tissue groups. Downsampling was applied, and 11,592 cells were included for each group. High relative cell density is shown as bright magma. j) Distribution of CD8 T cell states across tissue groups. Top bar plot showing the relative proportion of cells from 4 different tissue types for each CD8 T cell subset. Heatmap showing tissue prevalence estimated by Ro/e. k) Box plots showing cellular fractions of three CD8 T cell subsets across tissue groups. Each dot represents a sample. Pie charts displaying tissue composition. H, normal tissues from healthy donors; U, tumour adjacent uninvolved tissues; P, primary tumour tissues; and M, metastatic tumour tissues. The one-sided Games-Howell test was applied to calculate the p values between tissue types (sample number n = 20, 51, 156, 39), followed by FDR (false discovery rate) correction. FDR adjusted p-value: *≤0.05; **≤0.01; ***≤0.001, ****≤0.0001. For CD8_c1, p = 7.27e-10 (H vs. P), p = 4.21e-6 (H vs. M), p = 2.84e-4 (U vs. M), p = 8.38e-4 (P vs. M). Boxes, median ± interquartile range; whiskers, 1.5× interquartile range.
