Abstract
Brain tumors are one of the leading causes of cancer death; screening early is the best strategy to diagnose and treat brain tumors. Magnetic Resonance Imaging (MRI) is extensively utilized for brain tumor diagnosis; nevertheless, achieving improved accuracy and performance, a critical challenge in most of the previously reported automated medical diagnostics, is a complex problem. The study introduces the Dual Vision Transformer-DSUNET model, which incorporates feature fusion techniques to provide precise and efficient differentiation between brain tumors and other brain regions by leveraging multi-modal MRI data. The impetus for this study arises from the necessity of automating the segmentation process of brain tumors in medical imaging, a critical component in the realms of diagnosis and therapy strategy. The BRATS 2020 dataset is employed to tackle this issue, an extensively utilized dataset for segmenting brain tumors. This dataset encompasses multi-modal MRI images, including T1-weighted, T2-weighted, T1Gd (contrast-enhanced), and FLAIR modalities. The proposed model incorporates the dual vision idea to comprehensively capture the heterogeneous properties of brain tumors across several imaging modalities. Moreover, feature fusion techniques are implemented to augment the amalgamation of data originating from several modalities, enhancing the accuracy and dependability of tumor segmentation. The Dual Vision Transformer-DSUNET model's performance is evaluated using the Dice Coefficient as a prevalent metric for quantifying segmentation accuracy. The results obtained from the experiment exhibit remarkable performance, with Dice Coefficient values of 91.47 % for enhanced tumors, 92.38 % for core tumors, and 90.88 % for edema. The cumulative Dice score for the entirety of the classes is 91.29 %. In addition, the model has a high level of accuracy, roughly 99.93 %, which underscores its durability and efficacy in segmenting brain tumors. Experimental findings demonstrate the integrity of the suggested architecture, which has quickly improved the detection accuracy of many brain diseases.
Keywords: Dual vision transformer, Feature fusion, Brats dataset, Brain tumor segmentation, Dice coefficient
Abbreviations:
| Terms | Abbreviations | Terms | Abbreviations |
| NBTS | National Brain Tumor Society | HGG | high-grade gliomas |
| LGG | low-grade gliomas | MR | magnetic resonance |
| MRI | Magnetic resonance imaging | BRATS | Brain Tumor Segmentation |
| AI | Artificial Intelligence | CNN | Convolutional Neural Network |
| NLP | Natural Language Processing | DT | Dual Vision Transformer |
| FLAIR | Fluid-Attenuated Inversion Recovery | ViT | Vision Transformers |
| SDS-MSA-Net | The selectively deeply supervised multi-scale attention network | SDS | Selective Deep Supervision |
| WT | Whole Tumor | TC | Tumor Core |
| ET | Enhanced Tumor | GFI | global feature integration |
| RCNN | Region-based Convolutional Neural Networks | FHSA | Fusion-Head Self-Attention Mechanism |
| IDFTM | Infinite Deformable Fusion Transformer Module | T1Gd | T1-weighted |
| ED | Edema peritumoral | EDA | Exploratory Data Analysis |
| IoU | Intersection over Union |
1. Introduction
Brain tumors pose a substantial global health issue, as they have the potential to inflict extensive harm on the central nervous system [[1], [2], [3], [4]]. Neoplasms, defined by the aberrant and unregulated proliferation of neural cells within the cranial cavity, provide a substantial risk to human survival. According to the National Brain Tumor Society (NBTS), an estimated 90,000 individuals in the United States are affected by primary brain tumors annually, which include both malignant and non-malignant instances [5,6]. It is a matter of concern because an estimated annual mortality rate of approximately 20,000 individuals is attributed to brain tumors and other malignancies affecting the nervous system in the United States [7,8]. The aforementioned statistics demonstrate a consistent upward trend, underscoring the pressing necessity for developing and implementing sophisticated and efficient approaches to diagnosis and treatment. Gliomas, which are the most common primary brain tumors in adults, exert a particularly severe impact on the central nervous system [9,10]. Gliomas can be categorized into two primary classifications: low-grade gliomas (LGG) and high-grade gliomas (HGG). HGG, in particular, exhibits a more aggressive nature and is distinguished by its rapid growth. Individuals who receive a diagnosis of high-grade glioma (HGG) generally experience a life expectancy of two years or less [[11], [12], [13]]. This highlights the significant need for precise and prompt detection for appropriate treatment and care. Recently, there has been an increasing scholarly focus on advancing artificial intelligence (AI) methodologies for brain tumor segmentation. AI algorithms, namely deep learning networks, have demonstrated considerable potential in the field of automated brain tumor segmentation [14]These algorithms have exhibited promising outcomes by accurately identifying and outlining tumor boundaries. Consequently, they have proven valuable tools for healthcare practitioners in developing treatment plans for patients with brain tumors.
Brain tumors represent a substantial medical concern, giving rise to a multitude of health complications and presenting obstacles in terms of precise diagnosis and formulation of treatment strategies [15,16]. In recent times, there has been a notable increase in the number of research projects that concentrate on the segmentation of brain tumors to enhance the precision and effectiveness of tumor identification [[17], [18], [19]].
Developing and applying deep learning methodologies have significantly advanced computer vision endeavors, particularly in the healthcare domain, specifically in segmenting brain tumors. Deep learning techniques have transformed considerably multiple domains of image analysis and pattern recognition, showcasing remarkable efficacy across a diverse array of applications [[20], [21], [22]]. Deep learning (DL) techniques have demonstrated potential in the field of healthcare for addressing challenges associated with the interpretation of medical images, including the segmentation of brain tumors [[23], [24], [25]]. Specifically, these methodologies have facilitated the process of distinguishing several components of cerebral neoplasms, such as the entirety of the tumor (WT), the augmented tumor (ET), and the central region of the tumor (TC) [[26], [27], [28]].
Applying deep learning techniques has proven highly advantageous in segmenting brain tumors, enabling the identification and differentiation of various tumor components. However, the precise and reliable delineation of the increased and core tumor regions remains a challenging task [[29], [30], [31]]. The tumor portions exhibit distinct features such as reduced dimensions, asymmetrical contours, and comparable textures to the adjacent normal tissues. Due to these circumstances, the existing techniques employed for the segmentation of the ET and TC regions do not consistently yield the desired outcomes [[32], [33], [34]]. In light of the aforementioned constraints, the primary objective of this study is to introduce an innovative deep learning-driven framework that enhances the accuracy of segmenting both the increased and core tumor regions in brain tumors [[35], [36], [37]]. The proposed system seeks to leverage the capabilities of deep learning techniques to address the challenges associated with segmenting small, irregular tumor regions that exhibit textures like those of healthy tissues [38]. Our research endeavors to boost the accuracy of tumor region segmentation by utilizing a robust deep-learning architecture and implementing customized pre-processing approaches. This approach outperforms existing state-of-the-art methods to identify enhanced and core tumor regions accurately.
Further, it is essential to segment brain tumors as precisely as possible to diagnose the disease correctly, as well as to plan appropriate treatment and assess its efficacy. General approaches can poorly solve the task of segmenting small, irregular, and complex protuberant tumor regions and can negatively impact patient treatment. The Dual Vision Transformer-DSUNET with Feature Fusion for Brain Tumor Segmentation has been devised to overcome these significant limitations of existing methods. Moreover, the proposed approach uses improved dual vision transformers to improve the segmentation step by identifying fine details and adjusting the context of the imaging data from different modalities. This method believes that integrating feature fusion methods can eliminate the defects of single-modality and provide a more comprehensive understanding of tumor features. Improving segmenting in both core and enhanced areas places the diagnosis at a more accurate level and applies better clinical decision-making. Further, the novelty of this work is evident from the fact that the proposed method can be used to segment brain tumors, and it can provide a reliable solution to a complex problem in medical image analysis. Closing this gap is the potential of the proposed Dual Vision Transformer-DSUNET methodology for developing the neuroimaging field and improving the patients’ outcomes due to a more accurate definition of the tumor.
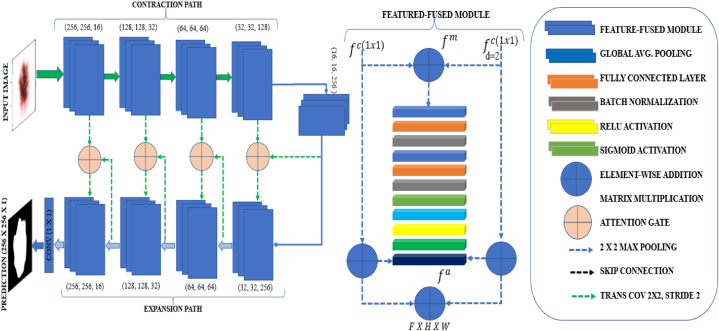
Fig. 1 depicts the framework of the dual vision transformer (DSUNET) with feature fusion, which is employed for brain tumor segmentation. The research employs the Brats 2020 dataset comprising T1ce, T2, T1, and FLAIR images. The data pre-processing procedures encompass several essential phases, such as data partitioning, intensity normalization, image resizing, gaussian sampling, smoothing, tumor label management, and NIFTI data manipulation. The images are subsequently inputted into the DSUNET, a dual-vision transformer incorporating feature fusion. The outcomes are assessed using diverse measures, including loss function, dice coefficient, blob loss, and IoU. The present system facilitates the identification of brain tumors by leveraging the analysis of captured images.
Fig. 1.
Framework for the dual vision Transformer-DSUNET with feature fusion for brain tumor segmentation.
The contributions of the research paper are as follows.
-
i.
The research presents a novel model called Dual Vision Transformer-DSUNET, which uses state-of-the-art transformer-based architectures in conjunction with a new DSUNET architecture. This model is planned to segment brain tumors without external input and with high accuracy using dual vision approaches that will capture all the characteristics of tumors in different types of images.
-
ii.
The study offers a novel contribution to feature fusion approaches, enhancing the possibility of combining information from different imaging modalities. This approach significantly enhances the accuracy and credibility of the segmentation of brain tumors by integrating various data sources to generate a precise analysis of tumor attributes.
-
iii.
The evaluation based on the Dice Coefficient shows that the presented Dual Vision Transformer-DSUNET model exhibits higher efficiency in tumor segmentation compared with existing approaches. The high Dice Coefficient scores are evident, and they signify the model's ability to demarcate the tumor region well enough for diagnosis and treatment planning.
-
iv.
The study increases segmentation accuracy by proposing new techniques to improve segmentation quality and applying multi-modal data to patients with brain tumors' diagnostic and therapeutic flow. Enhancing the segmentation functionality of the model results in its ability to provide clearer images that would enable better visualization of tumor morphology and growth patterns, which may enhance clinicians' clinical decisions and enable them to develop appropriate treatment plans.
-
v.
The study reveals the improvement of computational imaging methods with the help of dual vision transformers and feature fusion. This novel strategy also overcomes the limitations of current research in brain tumor segmentation and becomes the new standard for future improvement of medical image analysis methods.
This work explores brain tumor segmentation in various sections. Section II discusses current methods and their flaws, while Section III details experiment dataset collection and pre-processing. Section IV studies the Dual Vision Transformer-DSUNET with the Feature Fusion method, enhancing precision. Section V compares experiments to literature-reviewed methodologies and concludes that brain tumor segmentation improves diagnosis and treatment. Section VI describes the discussion part. Section VII of the research article discusses major findings, effects, and future directions. The article concludes brain tumor segmentation improves diagnosis and treatment.
2. Literature review
The segmentation of brain tumors plays a crucial role in facilitating the detection and treatment of medical conditions through imaging techniques [2,3,5]. The challenging job of segmenting brain tumors has been the subject of numerous approaches and algorithms.
Liu et al. [7] presented a study wherein they employed a pixel-level and feature-level image fusion approach to address the challenge of brain tumor segmentation in multimodal MRI, as discussed in their published work. The proposed technique was evaluated on 369 cases from the BraTS 2020 dataset. The researchers employed a methodology that entailed the creation of PIF-Net, a convolutional neural network (CNN) based 3D pixel-level image fusion network. The objective was to enhance the input modalities of the segmentation model. The methodology yielded positive outcomes, as evidenced by a dice coefficient of 0.8942 for the overall tumor and an average dice coefficient of 0.8265. This study exhibits notable limitations. As mentioned earlier, the methodology showed favorable performance on the BraTS dataset; however, its suitability for alternative datasets or real-time scenarios remains unexplored [8].
Moreover, the study conducted by Samosir et al. [9] sought to enhance the brain tumor segmentation process by combining diverse methodologies. The study employed a combination of CNN, the double-density dual-tree complex wavelet transform (DDDTCWT), and the genetic algorithm (GA) to partition brain MRI data accurately. The researchers employed the DDDT-CWT technique to partition the tumor images. Subsequently, a CNN was utilized for further analysis. A Genetic Algorithm (GA) was also employed for optimization, using 48 distinct combinations. A test dataset consisting of 913 images was utilized to get an F-1 score of 99.42 %. The results obtained were deemed satisfactory. The genetic algorithm was also employed to optimize the weights and biases of the initial layer of the CNN. The evaluation metric value of the dice coefficient exceeded 0.90 [[11], [12], [13]].
Likewise, in their study, Ottom et al. [17] Discuss using the 3D-Znet deep learning network to effectively separate 3D MR brain tumors. The model aims to optimize the representation of features at various hierarchical levels and achieve high performance by utilizing a variable autoencoder Znet and completely connected dense connections. Three-D-Znet consists of I/O blocks, four encoders, and four decoders. The encoder-decoder blocks consist of dual convolutional 3D layers, batch normalization, and activation functions [18,20,25]. The training and testing of this model were conducted using BraTS2020, a multimodal stereotactic neuroimaging dataset that includes tumor masks. Based on the dice coefficient metric, the pre-trained 3D-Znet model achieved scores of 0.91 for WT, 0.85 for TC, and 0.86 for ET. These ratings indicate 3D-Znet's brain tumor segmentation precision. This study's scope was limited to utilizing a single dataset for training and validation. The problem of segmenting medical 3D images, particularly brain tumors, was addressed by Jiang et al. [21] through the utilization of the Swin Transformer architecture. The researchers employed widely-used datasets, namely Brats 2019, 2020, and 2021, to train and assess the efficacy of their algorithm. The approach included a combination of Swin Transformer, convolutional neural networks, and encoder-decoders to enhance the semantic understanding derived from the Transformer module and improve feature extraction. The findings indicate that the SwinBTS model demonstrates proficiency in detecting tumor boundaries among intricate three-dimensional medical images. The entire tumor achieved a score of 82.5 %, whereas the score for the core was 69.85 %.
Moreover, BTS can identify and pinpoint medical imaging abnormalities to facilitate accurate diagnosis and effective treatment, according to the authors, Deepa et al. [23], propose a modification to the 2D-UNet architecture for brain tumor segmentation. The most recent iteration of the 2D-UNet architecture demonstrates the ability to process 2D and 3D data across several modalities. The segmentation map of the network, which is of equal dimensions as the input image, exhibits dice and intersection-over-union scores of 0.899 and 0.901, respectively. The use of statistical methods can accurately identify tumor regions. The enhanced 2D-UNet demonstrates superior performance compared to existing methodologies. The utilization of models and three-dimensional volumetric data enhances the analysis of brain tumors [24,26,27].Despite achieving success, it is essential to acknowledge that the process and model have inherent limitations. The segmentation process is influenced by factors such as the size, location, and structure of cancer. To apply and extend the approach, it is necessary to utilize more prominent and varied datasets and conduct thorough validation procedures.
In their study, Gai et al. [28] propose the utilization of a residual mix transformer encoder in conjunction with a convolutional neural network to distinguish between 2D brain tumors. A mix transformer in the system mitigates the loss of patch boundary data, enhancing patch embedding quality. The GFI module of the feature decoder incorporates global attention to improve contextual understanding. This study conducted a comprehensive investigation on the segmentation of brain tumors utilizing LGG, BraTS 2019, and BraTS 2020 datasets. The RMTF-Net has superior subjective and objective visual performance, surpassing global standards. The mean Dice and Intersection over Union (IoU) scores were calculated to be 0.821 and 0.743, respectively. The limited usefulness of studying solely 2D brain tumor segmentation is shown in its restricted generalizability to other medical image segment [29,30]The efficacy of the suggested technique may vary across different datasets, necessitating more investigation into its applicability to diverse populations.
Similarly, the scholarly paper by Nian et al. [32] presents a novel brain tumor segmentation strategy via a 3D transformer-based approach. The Fusion-Head Self-Attention Mechanism (FHSA) leverages the long-range spatial dependency of 3D MRI data. The self-attention module known as the Infinite Deformable Fusion Transformer Module (IDFTM) is designed to extract features from deformable feature maps in a plug-and-play manner [33,34]. The proposed technique outperformed leading segmentation algorithms on publicly accessible BRATS datasets. The Dice coefficients for the entire tumor, core tumor, and enhanced tumor were found to be 0.892, 0.85, and 0.806, respectively.
Furthermore, Zongren et al. [35] they proposed a method for the segmentation of brain tumors. Cross-windows and focused self-attention in the multi-view brain tumor segmentation model enhance global dependency and receptive field. The model's receiving field is expanded by parallelizing horizontal and vertical fringe self-attention in the cross window, increasing coverage without incurring additional computational costs. The concept of self-attention has been shown to effectively capture both short- and long-term visual dependencies, as demonstrated in previous studies [[36], [37], [38]].The model performed well in classifying core tumors, achieving an accuracy rate of 88.65 %. Additionally, it exhibited improved segmentation capabilities, with an accuracy rate of 89.39 %.
Several methods have been presented in recent approaches for segmenting brain tumors. Zhu et al. [39] used the Sparse Dynamic Volume TransUNet with multi-level edge fusion to improve tumor delineation using BRATS 2020; however, the model was limited by high computational requirements and data heterogeneity. Further, an Adaptive Cross Feature Fusion Network was proposed by Yue et al. [40], and it was proved to be helpful, although it demanded abundant training data and was apt to be overfit. Likewise, Liu et al. [41] put forward a multi-task model called SF-Net with outstanding fusion capacity but weak ability of generalization owing to its complicated structure. In addition, Berlin et al. [42] introduced an explainability-focused hybrid PA-NET with GCNN-ResNet50; while its architecture is quite complex, this work suffered from high computational complexity and scalability problems. On the same, Yue et al. [43] introduced the Adaptive Context Aggregation Network, and while it achieved generally favorable results, the efficacy was inconsistent over the different imaging protocols. However, the limitations as mentioned earlier are avoided in our proposed DVIT-DSUNET with the feature fusion technique, where we improve the accuracy of the geometric feature by combining dynamic volume information with the feature fusion technique to increase the model's accuracy and applicability while cutting the computational cost.
In this study, we introduce the Dual Vision Transformer-DSUNet, an innovative model for segmenting advanced brain tumors. This study's brain tumor segmentation model utilizes the robust Dual Vision Transformer and U-Net architecture to achieve high accuracy and computational efficiency. Our methodology was initially designed to incorporate dual attention. To perform tumor segmentation, we acquired both global and local contextual information. The proposed research addresses the challenge of accurately segmenting brain tumors using a novel dual-focus fixed scale and context approach. Our model employed distinct feature fusion methodologies. The utilization of dual-vision transformers and U-Net architecture has led to advancements in image segmentation. The integration of features enables the utilization of the respective strengths of many models while mitigating their limitations, enhancing the proposed model's efficacy. DSUNet outperformed the current leading methods on benchmark datasets. The segmentation accuracy of our newly developed brain tumor segmentation approach surpassed that of the dice similarity coefficient and intersection over the union, indicating a significant performance improvement. The medical image analysis of Dual Vision Transformer-DSUNet is enhanced by incorporating dual attention mechanisms and feature fusion techniques. This technique enables clinicians to identify and develop treatment plans, improving patient outcomes effectively. There is a need for further investigation and implementation of the notion above. Table 1 overviews previous sources, encompassing datasets, techniques, constraints, and outcomes.
Table 1.
List of Past References, including datasets/parameters, Approach, Limitations, and Results.
| Ref. | Datasets | Approach/Methodology | Limitations | Results |
|---|---|---|---|---|
| [14] |
|
Supervised multi-scale attention network (SDS-MSA-Net) |
|
Dice Coefficient for Whole Tumor is 90.24 %. |
| [17] |
|
3D-Znet, Convolutional Neural Network |
|
Dice Coefficient for whole tumor is 0.91, tomor core is 0.85 and enhanced tumor is 0.86. |
| [21] |
|
SwinBTS, Convolutional Neural Network |
|
SwinBTS dice score for whole tumor is 82.85 %, Core tumor is 69.85 % and the enhanced tumor is 59.67 %. |
| [23] |
|
BTS (Brain Tumor Segmentation), 2DU-Net. |
|
Dice Score of 0.899 and 0.90 for IoU. |
| [28] |
|
Residual Convolutional Neural Network (RCNN), Global Feature Integration (GFI), Residual Mix Transformer Fusion net. |
|
MeanDice and Mean IoU are 0.821 and 0.743. |
| [32] | It uses Brats dataset, with LGG. It contains two hundred and fifty-nine high grade Glimos, and seventy-six low grade gliomas. |
3D Brainformer, Fusion-head Self-Attention Mechanism, Infinite Deformable Fusion Transformer Module (IDFTM) |
|
Dice Coefficient for Whole tumor is 0.892, Core tumor is 0.85, and Enhanced Tumor is 0.806. |
| [35] | It uses Brats 2019 and Brats 2021 Datasets. | Convolutional Neural Network (CNN), PSwinBTS, Unet3D. |
|
Dice average for core and enhanced tumor are 88.65 % and 89.39 %. |
3. Data collection and processing
3.1. BRATS 2020 dataset
The BRATS (Brain Tumor Segmentation) challenge is an annual competition that focuses on the automated segmentation of brain tumors in magnetic resonance imaging (MRI) images [7,9,14,17]. Researchers may use the BRATS dataset to develop and evaluate new brain tumor segmentation methods.
The data set for BRATS 2020 is composed of:
Modalities of MRI.
-
•
Weighted by T1 (T1).
-
•
Contrast-enhanced T1-weighted imaging (T1ce).
-
•
Weighted by T2 (T2).
-
•
FLAIR stands for fluid-attenuated inversion recovery.
-
•
Mask (seg).
3.2. Ground truth labels
All of the BraTS multimodal scans are available as NIfTI files, which display the native (T1), post-contrast T1-weighted (T1Gd), and d) T2-FLAIR volumes. They were collected utilizing a range of clinical protocols and scanners from several (n = 19) institutions, which are acknowledged as data sources.
Each imaging dataset was manually segmented by one to four raters using the same annotation method, and knowledgeable neuroradiologists approved the annotations. The peritumoral edema (ED—label 2), the GD-enhancing tumor (ET—label 4), and the necrotic and non-enhancing tumor core (NCR/NET—label 1) are all noted.
Various tumor subregion annotations.
-
•
NCR/NET: Necrotic and non-enhancing tumor core
-
•
Edema peritumoral (ED)
-
•
(ET) GD-enhancing tumour
3.3. Data splits
-
•
Training data: The training set contains 70 % of the total data set, while the remaining 30 % is used to test the model. This subset consists of MRI images and ground truth annotations used in the models' training.
-
•
Data utilized for validation: In the presented dataset, 15 % is partitioned for validation. This subset includes MRI scans different from the training set and is used to adjust various model parameters and assess the latter's performance during the model-building process.
-
•
Test Data: The remaining 15 % is reserved for testing purposes. It is a set of MRIs used only to test the models' performance.
To design and test their brain tumor segmentation algorithms, researchers use this dataset to acquire accurate and reliable segmentation of tumor locations in brain MRI images.
3.4. Data pre-processing
The pre-processing techniques applied and how they affected the segmentation of brain tumors in the Brats 2020 data are detailed below.
-
1.
Normalization (Intensity Scaling): The MRI data's normalized voxel intensities range from [0, 1]. Normalization standardizes the intensity levels, allowing accurate training-related convergence and reliable comparisons.
-
2.
Resizing: MRI images are scaled using a preset dimension (IMG_SIZE by IMG_SIZE).
Resizing keeps the input's dimensions uniform, enabling the data to be used with the model and streamlining computations.
-
3.
Gaussian Smoothing: The MRI images are given Gaussian blur using a 3x3 kernel. Smoothing makes the data smoother and may enhance feature extraction by reducing noise and presenting more precise information.
-
4.
One-Hot Encoding for Masks: The different tumor classifications are represented by segmentation masks that have undergone one-hot encoding. Through one-hot encoding, the original label-based segmentation is transformed into a format suitable for training, aiding the model's understanding of class links.
-
5.
Handling Tumor Labels: The numerous classes of tumors are represented by segmentation masks that have been one-hot encoded. One-hot encoding transforms the initial label-based segmentation into a format appropriate for training, aiding the model's understanding of class links.
-
6.
Intensity Normalization for NIFITI Data: The NIFTI data is adjusted to ensure homogeneous intensity ranges. To maintain consistent intensity scales throughout the collection, the NIFTI data should be standardized.
Before processing the MRI data, these procedures ensure that it is smooth, noise-free, and suitable for applying the Dual Vision Transformer-DSUNET model for effective segmentation and training. The segmentation is improved, and the model is more accurate with the help of the normalized and preprocessed data, resulting in a more trustworthy brain tumor segmentation model.
3.5. Exploratory data analysis
To acquire insight into the brain tumor segmentation dataset, we will undertake exploratory data analysis (EDA) in this part utilizing a variety of visualizations. The ROI overlay graphic highlights the Flair modality's particular region of interest. The colored overlay represents the segmented tumor region. With the help of the slice viewer, each modality (Flair, T1, T1CE, and T2) can explore various slices. It enables us to look at the differences in tumor structures between slices. The distribution of pixel values in the Flair modality is displayed in the histogram representation. For image processing tasks, it offers a comprehension of the intensity distribution.
The heatmap visualization highlights the intensity distribution in a 2D heat map by displaying a slice of the Flair modality. This aids in comprehending how pixel intensities vary spatially. The Flair modality's structure outlines are seen on the contour plot. It gives the tumor boundaries a visual representation, making comprehending its size and shape easier. The comparative visualization permits comparing numerous patients' images from various modalities (Flair, T1, T1CE, and T2). It enables us to spot differences in tumor features between patients and treatment regimens.
The contour map shows the structural outlines of the Flair modality. It visually represents the tumor's borders, making understanding its size and shape more straightforward. Comparative visualization allows us to compare images of multiple patients obtained using different modalities (Flair, T1, T1CE, and T2). It enables us to identify variations in tumor characteristics between patients and therapeutic approaches.
3.6. Region of interest (ROI) overlay
The image (Fig. 2) displays a well-separated brain tumor in a particular slice (index 80) of the Flair modality. The overlay effectively draws attention to the region of interest inside the segmented tumor area. Color mapping generates a sharp visual contrast that can be used to distinguish and assess the tumor's geographic spread and borders.
Fig. 2.
Region of interest overlay for Fair, t1, t2, t1ce, and Mask Modality.
Before creating a binary mask to represent the tumor location, image data must be transformed to uint8 for viewing to generate the overlay. This mask is then converted into a colorful overlay using a Jet colormap. By combining the Flair modality image and the colored mask, the final overlay achieves harmony between the initial grayscale image and the highlighted tumor spot. The analytical procedure for segmenting brain tumors depends heavily on this image, which is necessary for understanding the tumor's location. This picture helps us understand the distinctions between different modalities' areas of interest.
3.7. Slice viewer
The provided visualization (Fig. 3) has a unique slice viewer tool for displaying slices of various picture modalities. Each slice can be accessed using keyboard shortcuts ('j' for the prior slice and 'k' for the following slice). This interactive viewer offers a rapid method for examining the spatial characteristics of each modality, aiding in comprehension of the comparison of the brain images.
Fig. 3.
Brain tumor slices for Each Modality.
The function displays the central slice of the supplied picture data while initializing a subplot layout. Moving between the slices causes a dynamic updating of the visible slice index. The 'process_key', 'previous_slice', and 'next_slice' functions enable smooth slice navigation, enabling in-depth examination of each modality. The program is then applied to every picture modality (Flair, T1, T1CE, and T2) for a complete dataset analysis and to better understand each modality's unique features. A single modality may have several slices; in this case, we studied a certain number of slices, and with the use of visualization, we discovered how each modality is interpreted as a slice.
3.8. Histogram visualization
Histogram visualization, as shown in Fig. 4, is necessary to comprehend the distribution of pixel intensities within a specific picture modality. The “function,” in this case, flattens the image data into a 1D array. The frequency of each pixel value is then plotted on the y-axis of the histogram versus the total number of occurrences. The histogram helps us comprehend the intensity distribution by emphasizing potential peaks and patterns.
Fig. 4.
Histogram of Intensity Distribution of Flair, t1, t2, t1ce, and Maks Modalities.
This image demonstrates that the intensity distribution of almost all modalities is comparable, with the peak intensity being the same and other smaller intensities having slightly different values. Identifying the typical intensity values of various tissues and anomalies in brain imaging aids in further analysis and feature extraction. As an illustration, the Flair modality may show peaks corresponding to different brain tissues and anomalies, providing details about the characteristics of the brain tumor region.
3.9. Heatmap visualization
Heatmaps, as shown in Fig. 5, are pixel intensities for a specific slice of an imaging modality. They help highlight patterns and data variations. The program generates a heatmap with various intensities represented by various colors using the Seaborn library. We'll use slice 80 for this illustration.
Fig. 5.
Heatmap of Flair, t1, t2, t1ce and Mask Modality.
3.10. Contour plot
Contour plots in Fig. 6 show intensity or structural differences and can be used to draw boundaries or highlight certain regions of interest within an image. The technique creates a contour plot centered on a particular slice of the provided image data (in this case, slice 80) using the matplotlib library. Contours are shown with a linewidth of 0.5 and are rendered in red ('r') for better visibility.
Fig. 6.
Contour Plot of Flair, t1, t2, tice and Mask Modality.
Contour charts are handy for identifying distinct structures or anomalies based on intensity changes. This brain imaging visualization technique can assist in locating specific regions or issues in the brain slice by providing a visual pathway for future investigation. By displaying outlines on a particular slice, we may better understand the picture's organizational structure and identify potential brain regions of interest.
3.11. Comparative visualization
Medical imaging frequently uses a variety of imaging modalities for the same patient. The feature makes it easier to compare various modalities effectively. With individual patients in each row and different imaging modalities (such as Flair, T1, T1CE, and T2) in each column, it generates a grid of images, as shown in Fig. 7. By displaying these images, clinicians and researchers can visually compare how different modalities depict specific parts of the brain's architecture or pathology. To create models that effectively combine the data from all modalities for more accurate segmentation or analysis of brain tumors, understanding each modality's advantages and disadvantages can be crucial. The technique also ensures that each subplot has a unique name, making it easy to identify the modality and patient connected to each modality.
Fig. 7.
Comparative modalities of two patients.
To develop a segmentation model that will be effective for assessing brain tumors, we must have a deeper understanding of the dataset's characteristics and the locations of the tumors. By emphasizing the segmented tumor zone, the overlay aids in identifying the particular region of focus in the Flair modality. It is possible to identify differences in tumor architecture by comparing various slices. Understanding these variations is essential for effective segmentation. The histogram displays the distribution of pixel values. The distribution of pixel intensities can be inferred from the fact that most pixel values fall within a specific range. By giving an impression of the intensity fluctuation across the slice, the heatmap aids in detecting locations with high or low intensities.
The contour plot depicts the tumor's structure, making it more straightforward to identify the tumor's borders and overall shape. We can observe changes in tumor characteristics by comparing different modalities for different patients. This comparison is crucial for comprehending the differences and overlaps between tumor representations in various imaging modalities.
4. Model design
The critical task of segmenting brain tumors in medical imaging significantly impacts patient care, treatment planning, and diagnosis. The precise and thorough definition of brain tumor regions is necessary for medical practitioners to make educated judgments. Using multi-modal MRI data from the BRATS2020 dataset, we introduce the Dual Vision Transformer-DSUNET model with feature fusion, a novel method for classifying brain cancers.
Modern cutting-edge models for brain tumor segmentation usually have trouble effectively utilizing spatial features and extensive contextual data. Transformers have demonstrated potential in several sectors and excel at global context modeling. Similarly, it has been shown that the U-Net design effectively preserves spatial information throughout segmentation tasks. Due to the complementing qualities of these architectures, we present the Dual Vision Transformer-DSUNET model to efficiently integrate the global context modeling capabilities of transformers and the spatial preservation capability of U-Net for accurate brain tumor segmentation.
The DVIT-DSUNET starts with a DVIT encoder that uses the Transformer's self-attention mechanism to capture long-range dependencies and linkages within the input data. The DVIT encoder uses self-attention to convert an input tensor X:
| (1) |
Here, the function SelfAttention(X) applies the self-attention mechanism to the input tensor X, capturing global and local interactions between various X elements.
Dual stream architecture, used by DSUNET, combines two streams with independent attention techniques to offer improved feature representations. The output of the model is the union of these two streams:
| (2) |
To concentrate on crucial channels and geographical areas, the DSUNET model employs a dual attention mechanism. Let S represent spatial attention, and C represent channel-wise attention. The model combines these attentions to produce the final attention A:
| (3) |
In this case, ⊙ stands for element-wise multiplication.
Feature fusion combines features from several levels or sources to provide a complete picture. Let 1 F1 and 2 F2 represent characteristics from various sources. It is possible to express the feature fusion procedure as a weighted sum:
| (4) |
- where α and β are weights for each feature's contribution.
These equations represent the basic mathematical operations of the DVIT-DSUNET model, which combines components from DVIT, DSUNET, dual attention processes, and feature fusion to improve brain tumor segmentation performance.
Assume that the visual feature has a dimension of R PxC, where P is the number of patches, and C is the number of channels. The complexity is when the conventional global self-attention is used alone. It's outlined as:
| (5) |
- where head = Attention (Qi, Ki, Vi)where Xi stands for the ith head of the input feature, and Wi stands for the ith head's projection weights for Q, K, V, and
| (6) |
And:
| (7) |
and are dimensional visual features with heads.
It is important to note that the output projection WO has been left out of this. The computing cost is excessive given that P may be extremely big, such as 128,128.
window attention computes self-attention inside local windows. The windows are set up such that they uniformly and non-overlappingly divide the image. Assume that there are Nw distinct windows, each of which contains Pw patches, and that consequently, window attention might be described as
| (8) |
where the local window queries, keys, and values are Qi, Ki, Vi, and R Pw∗Ch, respectively. Window-based self-attention has an O(2P PwC+4P C2) computational complexity with a linear complexity with the spatial dimension P. Window attention can no longer simulate global information despite the decreased computation. We'll demonstrate how our suggested channel attention naturally resolves this issue and strengthens window attention.
We divide the channels into many groups and execute self-attention inside each group to minimize computational complexity. We have C = Ng Cg, where Ng is the number of groups and Cg is the number of channels in each group. By communicating across a group of channels with image-level tokens, we achieve global channel group attention.
It's outlined as:
| (9) |
Where Qi, Ki, Vi, and R PxCg represent a channel-wise grouping of image-level queries, keys, and values. we should be aware that even when we transpose the tokens in the channel attention, the projection layers W.
4.1. Objectives
Following are our two key objectives for this study.
-
•
Utilizing the benefits of transformers to extract global contextual information and distant linkages from multi-modal MRI data.
-
•
To enhance segmentation performance, particularly for regions of brain tumors with necrotic and non-enhancing tumor core (NCR/NET), peritumoral edema (ED), and GD-enhancing tumor (ET), by utilizing U-Net's capacity to preserve space and combining it with transformers' properties.
Fig. 8 is related to the distribution of DSUNET architecture layers, where the Dual Vision Transformer-DSUNET concept is offered with a dual-stream design. Each stream independently processes MRI data from multiple modes. Including transformer blocks in each stream facilitates the description of the overall environment, and the subsequent fusion of transformer-based characteristics with the U-Net architecture ensures the preservation of spatial information. Feature fusion, an important part of our strategy, improves the representation of the features obtained from the transformer and U-Net components.
Fig. 8.
DSUNET architecture layers Distribution.
This study advances the field of brain tumor segmentation by introducing a new model that combines the best features of U-Net and transformer-based topologies. The feature fusion component is crucial for fusing the spatial information preservation capabilities of U-Net with the global context awareness of transformers. Our state-of-the-art technique can significantly improve the accuracy and dependability of brain tumor segmentation, ultimately leading to superior therapeutic results.
4.2. Model architecture: dual Vision-DSUNET with feature fusion
A new architecture, Dual Vision-DSUNET with Feature Fusion, was developed to improve brain tumor segmentation in multi-modal MRI data in Fig. 9. This architecture uses dual attention approaches and feature fusion to improve segmentation accuracy by fusing the best elements of transformers and U-Net.
-
1.
Input Layer: The input layer accepts multi-modal MRI data, a crucial component of brain imaging for tumor segmentation.
-
2.
Encoder: The encoder component performs hierarchical processing to extract more complex properties from the input data. Max-pooling and ReLU activation come after convolutional layers for spatial reduction. The encoder continually learns new features, beginning with easier features and progressing to more intricate abstractions.
-
3.
Dual Attention Block: The architecture includes the dual attention block, a significant innovation. It employs two distinct attentional techniques: channel attention, which focuses on important channels by learning channel-specific weights, and spatial attention, which gives the feature maps' important spatial regions the necessary spatial attention. These attentions are coupled to enhance the feature representation for additional processing.
-
4.
Middle (Dual Attention): The intermediate component bridges the encoder and decoder. The feature representation is improved by using the dual attention block to capture local and global contexts.
-
5.
Decoder: The decoder component aims to reconstruct the original input resolution. It employs upsampling methods and the dual attention fusion block for richer feature integration. The decoder gradually improves the segmentation masks by including characteristics from several layers.
-
6.
Dual Attention Fusion Block: This block encourages collaboration between streams by enabling the combination of features from many network nodes. Combining dual attention ensures that the model gathers the most relevant input for segmentation.
-
7.
Output Layer: The final output layer can predict the segmentation masks for the subregions of brain tumors. A softmax activation function creates probability distributions across the segmentation classes. The model will distinguish several tumor subregions, including the necrotic and non-enhancing core, the peritumoral edema, the enhancing tumor, and the backdrop. To strike a compromise between employing transformers to obtain global context and using U-Net architecture to preserve space, the Dual Vision-DSUNET architecture was thoughtfully developed. The model gains a deeper understanding of the many components of the brain tumor thanks to the dual attention and fusion methods, which produce precise and trustworthy segmentation findings. This novel approach could profoundly impact the study of brain tumors and medical imaging.
Fig. 9.

DVIT-DSUNT layers blocks distribution.
4.3. Model blocks and layers distribution and properties
4.3.1 Dual Attention Block: The “Dual Attention Block” is a critical component that enhances the model's ability to focus on important information in both channel-wise and spatial dimensions. Below is a detailed explanation of this block.
-
a)
Channel Attention: The input feature map (x) is subjected to a 1x1 convolutional layer to drastically reduce the number of channels (usually ).
A ReLU activation is then performed to add non-linearity.
The features are converted to a sigmoid activation using a further 1x1 convolutional layer, which yields a channel attention map. Fig. 10 is for Channel Attention in DVIT.
Fig. 10.
Channel attention in DVIT.
Depending on their significance in the channel-wise dimension, individual elements are either emphasized or suppressed using the channel attention map. Here Fig. 11 shows the DVIT dual attention block architecture layers distribution.
-
b)
Spatial Attention
Fig. 11.
Dvit dual attention block Architecture layers distribution.
The input feature map (x) is immediately applied to another 1x1 convolutional layer with a sigmoid activation to produce a spatial attention map.
This spatial attention map in Fig. 12 emphasizes key geographic areas.
-
c)
Applying Attention
Fig. 12.
Spatial attention for DVIT dual attention.
The original input feature map (x) is element-wise multiplied by the channel- and spatial-specific attention maps. Consequently, a new feature map combines the original characteristics in a weighted manner, emphasizing particular channels and geographical areas according to the attention ratings.
4.4. Dual attention block
The “Dual Attention Fusion Block” achieves dual attention fusion by combining the outputs from two different routes (streams). A thorough description of this block is given below.
-
a)
Input: Two feature maps (x1 and x2) are provided as inputs to the block, often from various locations within the network.
-
b)
Fusion: A fused feature map (fused_features) is produced by joining the features from the two streams along the channel dimension, as shown in Fig. 13.
-
c)
Dual Attention: The fused features (fused_features) are given the dual_attention_block function to include dual attention. Both channel- and space-based attention processes are involved in this.
-
d)
Output: The output of the block, which symbolizes the fusion of features with dual attention, is the feature map that results from the dual attention process. The “Dual Attention Fusion Block” enables the model to synergistically merge features from various network nodes using the dual attention mechanism to use both spatial and global contexts. This fusion technique helps to accurately partition brain tumors and improves the model's capacity to collect critical characteristics. By concentrating the model's learning on pertinent spatial and channel-wise information, these attention methods are essential for enhancing segmentation performance when brain tumors are being segmented using the given DSUNET architecture.
Fig. 13.
Feature fusion operation for DVIT-DSUNET.
The parameters here show in Table 2, the training variables which are obtained after fine tuning the model for various runs. At these parameters the model performance is optimum leading to the less training time, higher performance and less computational Cost.
Table 2.
Model parameters.
| Parameters | Value |
|---|---|
| Learning Rate | 0.001 |
| Epochs | 50 |
| Batch size | 64 |
| Training Parameters | 2.1 M |
| Number of layers | 67 |
The other parameters being set are layers units.
The inital layer or first layer takes images of the sizes 128 by 128 as follows.
Shape of input: (128, 128, 2)
The details of these parameters is as given below in layers distribution.
The dual attention layer is being used as a block with set parameters of different numbers of neurons at different layers. All of the details are given below.
4.5. Layer distribution
-
a)
Input Layer
Shape of input: (128, 128, 2)
MRI data from many modes are fed into the model as shown in Fig. 14.
-
b)Encoder
-
-Conv 1: Convolutional layer with 32 filters, a 3x3 kernel, ReLU activation, and the same padding.
-
-Conv 2: Convolutional layer with 32 filters, 3x3 kernel, ReLU activation, and same padding.
-
-Pool 1: Max-pooling layer with a (2, 2) pool size.
-
-A convolutional layer with 64 filters, a 3x3 kernel, ReLU activation, and the same padding is called Conv 3.
-
-Conv 4: Convolutional layer with 64 filters, 3x3 kernel, ReLU activation, and same padding.
-
-Pool 2: Maximum pooling layer with a (2, 2) pool size.
-
-Conv5: A convolutional layer with 128 filters, a 3x3 kernel, ReLU activation, and the same padding is called Conv5.
-
-Conv 6: Convolutional layer with 128 filters, 3x3 kernel, ReLU activation, and some padding.
-
-Pool 3: Max-pooling layer with a (2, 2) pool size.
-
-
-
c)
Middle (Dual Attention)
Fig. 14.
Proposed model layers distribution.
A convolutional layer with 256 filters, a 3x3 kernel, ReLU activation, and the same padding is called Conv 7. A convolutional layer with 256 filters, a 3x3 kernel, ReLU activation, and the same padding is called Conv 8. Dual Attention Block: The characteristics of the middle layer are subject to a dual attention mechanism.
-
d)Decoder
-
-Up3: Up-sampling that recovers spatial information by employing nearest-neighbor interpolation (UpSampling2D).
-
-Fusion employing dual attention: Combines features from the second and third encoder layers.
-
-Conv9: A convolutional layer with 128 filters, a 3x3 kernel, ReLU activation, and' padding is called Conv9.
-
-Conv 10: A convolutional layer with 128 filters, a 3x3 kernel, ReLU activation, and the same padding is called Conv10.
-
-Up2: Nearest-neighbor interpolation used in up-sampling (UpSampling2D).
-
-Fusion employing dual attention: Combines features from the decoder and the second encoder layer.
-
-Conv 11: A convolutional layer with 64 filters, a 3x3 kernel, ReLU activation, and the same padding is called Conv11.
-
-Conv 12: A convolutional layer with 64 filters, a 3x3 kernel, ReLU activation, and' padding is called Conv12.
-
-Up1: Nearest-neighbor interpolation-based upsampling (UpSampling2D).
-
-Dual Attention Fusion: Fusion uses dual attention that combines the first encoder layer's and the decoder's characteristics.
-
-
The convolutional layer with 32 filters, a 3x3 kernel, ReLU activation, and the same padding is called Conv 13. The convolutional layer with 32 filters, a 3x3 kernel, ReLU activation, and the same padding is called Conv 14.
-
e)
Output Layer
Convolutional layer Conv 15: Softmax activation for segmentation masks, 4 filters, and a 1x1 kernel.
The Dual Vision-DSUNET model effectively utilizes global context modeling from transformers and spatial preservation from the U-Net architecture. The dual attention blocks and feature fusion mechanisms enhance the segmentation performance for brain tumor subregions using the BRATS2020 dataset.
The model is instantiated using the provided function create_ds_unet3p and input shape (128, 128, 2), creating a complete model ready for training and evaluation.
5. Methodology stage
5.1. Pre-processing and data preparation
-
a)
Modalities fusion: Effectively combining data from many magnetic resonance imaging modalities.
By ensuring uniformity and comparability throughout the dataset, normalization, and standardization.
Data augmentation: The use of methods to enlarge the training dataset and enhance model generalization, such as rotation, flipping, scaling, and translation.
5.2. Train validation and test split
For training, fine-tuning, and evaluating models, the dataset was divided into training, validation, and test sets.
Fig. 15 shows the distribution of images for train, validation, and test set for the brats2020 Dataset.
Fig. 15.
Train Test validation split.
5.3. Architecture and model design
The dual Vision Transformer-DSUNET model architecture is described in detail below. It includes components for the encoder, middle, and decoder, transformer block and U-Net architecture integration, and blocks with dual attention and fusion.
-
a)
Loss Function: The segmentation model is optimized by choosing a suitable loss function, such as categorical cross-entropy, focused, or blob loss.
-
b)
Training and optimization: Choosing an optimizer (like Adam), and scheduling the learning rate. Train the model on the preprocessed dataset using the chosen loss function and optimizer.
early termination to avoid overfitting.
-
c)
Metrics: Explain assessment metrics such as the Hausdorff distance, sensitivity, and specificity. Visualize model predictions in comparison to ground truth annotations—qualitative evaluation. Quantitative evaluation is calculated using assessment metrics on the test set to determine the model's performance.
5.4. Technical details of dual vision Transformer-DSUNET
The proposed model is the Dual Vision Transformer-DSUNET (DVIT-DSUNET), which is an improvement of the current state-of-art in brain tumor segmentation through integrating transformer and U-Net by performing exceptionally well in segmenting multi-modal Brain MRI data from the BRATS2020 dataset. This novel approach responds to two essential problems of segmentation methods: how to incorporate the global contextual information and simultaneously avoid loss of spatial details critical for tumor border definition.
The central unit of the DVIT-DSUNET is the so-called Dual Vision Transformer (DVIT) encoder, which is based on the transformer's ability to attend to regions that are far apart from each other and, thus, capture the global context inherent in the input MRI data. In particular, it allows the encoder to capture detailed interactions across the whole image, in contrast to the standard convolutional approaches that fail to incorporate the global context information into the process.
Adding to the DVIT encoder, DSUNET has another attentiveness system that works simultaneously at the channel and spatial levels. This block is designed to pay dual attention to highlighting the essential features and areas in the data to enhance the feature mapping. The channel attention is achieved by using the 1x1 convolution with the ReLU activation followed by the sigmoid activation so that the channel attention map can scale the critical channels. Spatial attention is also used to precisely guide attention to spatial locations of interest, ensuring that the model attends to the right places in the feature maps.
The architecture's novelty lies in the fusion of features typical of the DVIT encoder that models the global context of the input data with U-Net, which preserves spatial information. Further, the integration mechanism enables the model to combine features efficiently, and the feature enrichments enhance segmentation performances. For the decoder phase, DVIT-DSUNET uses upsampling to improve the spatial dimension and dual attention fusion blocks to merge features from different network layers. This approach makes it possible to incorporate detailed features at fine and coarse scales, allowing for accurate delineation of some of the sub-regions of the brain tumor, such as necrotic and non-enhancing cores, peritumoral edema, and enhancing tumors.
The architectural layout has several layers and blocks to enhance the system's performance. The encoder takes input through several convolutional and pooling layers, and the middle part has dual attention blocks for enhancing feature representation. The decoder then reconstructs the image resolution and employs dual attention fusion to achieve feature integration and improve segmentation precision. The network's last layer employs softmax activation to produce a probability distribution of the tumor subregions.
In general, the DVIT-DSUNET model has been distinguished by combining transformers' global context and U-Net's mechanism to preserve spatial details. It enhances segmentation accuracy and reliability by utilizing a two-set attention mechanism and state-of-the-art features fusion. This model constitutes a significant improvement in medical imaging; it helps improve the diagnosis and treatment of brain tumors.
6. Results
6.1. Model training and evaluation performance
This section assesses the effectiveness of the Dual Vision Transformer-DSUNET model for segmenting brain tumors. We use a variety of assessment criteria to thoroughly evaluate the model's accuracy, precision, sensitivity, specificity, and segmentation quality.
-
a)
Loss Function (Focal Loss): A specialized loss function called focal loss was created to address class imbalance in binary classification problems. It emphasizes complex samples while down-weighting straightforward data to improve model training.
| (10) |
Where pt is the balancing factor, α t is the focusing parameter, and) γ is the expected probability.
-
b)
Accuracy: The percentage of correctly categorized samples among all samples is how accuracy is calculated.
| (11) |
-
c)
Mean Intersection Over Union (Mean IOU): The percentage of samples that were properly identified out of all samples is how accuracy is calculated.
| (12) |
-
d)
Dice Coefficient: The similarity between the expected and actual segmentation masks is calculated using the dice coefficient.
| (13) |
-
e)
Precision: The precision determines what percentage of the overall positive forecasts are real positive predictions.
| (14) |
-
f)
Sensitivity (Recall): Sensitivity quantifies the percentage of actual positive outcomes that were predicted to be true positives.
| (15) |
-
g)
Specificity: Out of the real negatives, specificity determines the percentage of accurate negative forecasts.
| (16) |
-
h)
Blob Loss: A loss function used in picture segmentation tasks is called the Blob Loss. It seeks to penalize the model for producing unconnected or isolated segmentation “blobs”. Particularly in medical imaging, where continuous and coherent segmentation is crucial, these unconnected components frequently result in erroneous segmentations.
| (17) |
Where; : The number of instances (blobs) in the segmentation of the ground truth is num_instances. The expected segmentation mask for the ith occurrence is anticipated as predicted, y predicted, i. true, y true, i is the ith instance's ground truth segmentation mask. The dice coefficient is. The Dice coefficient, which compares the similarity of two segmentation masks, is a similarity metric between sets. Its value is (A, B). In this instance, it's utilized to gauge how well the anticipated and actual blobs line up. The Blob Loss penalizes the model for producing unconnected or fragmented segmentations by computing the Dice coefficient for each isolated instance in the predicted and genuine segmentations and averaging them to produce the final loss. Here Fig. 16 shows the graph for accuracy and loss performance.
Fig. 16.
Accuracy and loss performances.
Table 3 shows the model evaluation results for EDEMA, core, enhanced and All classes.
Table 3.
Model evaluation results.
| Evaluation Metric | Enhancing | EDEMA | Core | All Classes |
|---|---|---|---|---|
| Training Accuracy | 99.68 | 99.84 | 99.79 | 99.93 |
| Loss- Focal Loss | 0.1724 | 0.1434 | 0.1547 | 0.1621 |
| Mean IOU | 0.1421 | 0.1344 | 0.1547 | 0.1453 |
| Dice Coefficient | 91.47 | 90.88 | 92.39 | 91.29 |
| Dice Loss | 0.0912 | 0.0812 | 0.0714 | 0.0871 |
| Precision | 99.01 | 98.45 | 98.77 | 98.12 |
| Sensitivity | 96.54 | 98.25 | 97.29 | 97.45 |
| Specificity | 99.12 | 99.48 | 99.23 | 99.95 |
| Blob Loss | 1.5571e-09 | 1.7728e-09 | 1.8871e-09 | 1.2231e-09 |
The evaluation metrics collected provide crucial information about the potency and dependability of our system for segmenting brain tumors. As shown by its outstanding training accuracy of 99.93 %, our model has learned and generalized successfully from the training dataset, capturing complex patterns and characteristics relevant to brain tumors. Additionally, it achieves a low loss value (focal loss of 0.1621), which shows how well our model works to minimize training-related errors. The mean Intersection over Union (IOU) score of 0.1453, which shows a decent overlap between the predicted and ground truth segmentations, demonstrates the model's ability to detect tumor locations properly. A remarkable Dice Coefficient of 91.29 %, which shows a high degree of similarity between the predicted and actual segmentations, further supports the resilience and accuracy of our model. The evaluation metrics are displayed for all classes of combined prediction as well as for the prediction of various tumor types, suggesting that the model can accurately determine whether or not the patient has a tumor in the first place as well as which tumor type is present since some patients can have all three types or even just one class. Additionally, some people likely have tumor segmentation that is more of an augmenting kind than a core type. The model can accurately predict both the total number of classes and the individual classes of tumors.
The model's accuracy, sensitivity, and specificity values of 98.12 %, 97.45 %, and 99.95 %, respectively, illustrate its ability to accurately detect actual positive cases and decrease false positives and negatives. The meager blob loss value (1.2231e-09) indicates that the model effectively reduces fragmented or disconnected segmentations, which is necessary for precise medical imaging interpretation. Overall, these findings point to a robust, reliable model that can accurately classify brain cancers. More validation utilizing a range of datasets and thorough clinical testing is required to ensure the model's applicability and efficacy in real-world healthcare scenarios.
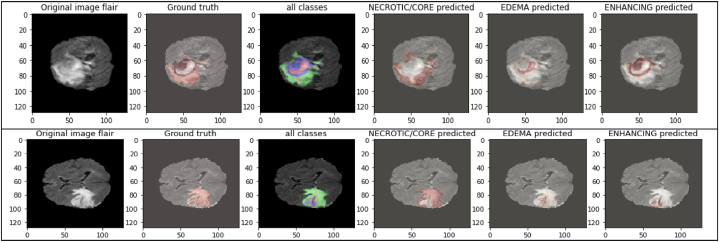
The images above in Fig. 17 display the images' immediate surroundings, which is the mask we want to conceal. The high training accuracy that was attained and the significant values for other assessment metrics, such as accuracy, mean IoU, precision, and sensitivity, as well as the correct predictions as shown in the above figure, demonstrate the impressive performance of the Dual Vision Transformer-DSUNET model for segmenting brain tumors. The model, which accurately captures tumor characteristics and delineates tumor locations, is a possible medical imaging technique for the research of brain tumors (see Fig. 18).
Fig. 17.
Prediction results for different modalities of masks.
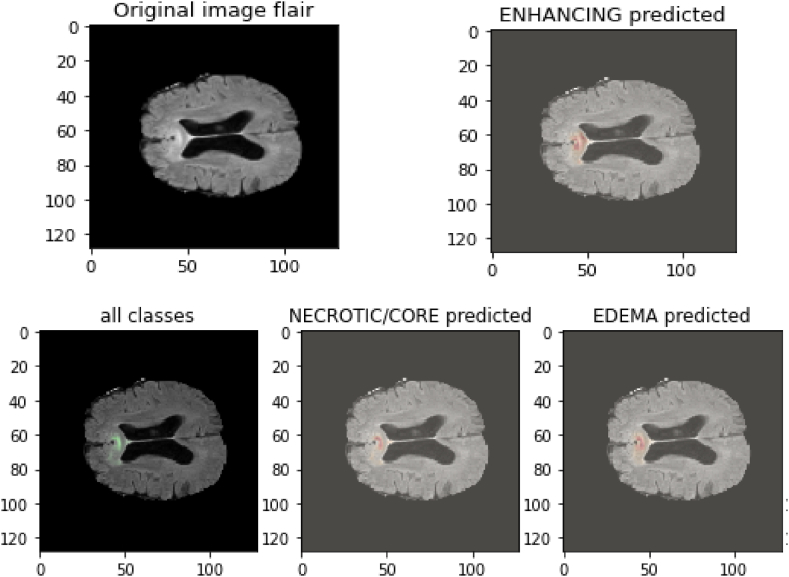
Fig. 18.
Predictions on Test images Flair of Different tumor types.
The results below display the whole brain scan, the type of tumor, and how our model identified it.
The flair-type image is used in the data above to forecast the many classifications of tumors we have, including core, edema, and enhancement. Since images cannot be given directly through our model, we convert all images into array types. The images shown above were chosen during model prediction and represent all image classes that our model can predict. It's also vital to note that only the flair images are included above because only flair images include a full scan of the brain tumor and all known tumor classes.
This part on model evaluation demonstrates in great detail how the Dual Vision Transformer-DSUNET model is excellent at precisely segmenting brain tumors. This demonstrates how it may be applied to aid physicians in identifying and caring for patients with brain disorders.
6.2. Model evaluation performance on Brats21 dataset
We assessed our model's performance using samples from the Brats21 dataset to determine its dependability and broaden its applicability to various brain MPI datasets that are not trained and are composed of multiple patients with different characteristics. The tumor class for samples in the dataset is predicted using the trained model on the Brats21 dataset. This way, we can determine whether our model is overfitting and performs admirably when tested against a different subset of the dataset. Additionally, the data would be accessible to any patient if our model were to be used in the medical area, making it crucial to test the efficiency and dependability of our model on various datasets. The following are some examples of Brats21 images, as shown in Fig. 19:
Fig. 19.
Brats21 Different Modalities samples images.
These images' forms are comparable to those in the Brats 2020 collection (155, 240, 240). We have four kinds of tumors: complete tumor, augmenting tumor, core tumor, and no tumor. These images are converted into array types with a generator and sent via our trained model prediction function. Following are the predicted classes of tumors as shown in Fig. 20:
Fig. 20.
Brats21 Predicted classes of Sample image 1.
In the places where the tumor is present, the images above in Fig. 21 demonstrate the projected classifications of tumor regions of various types, including augmenting, edema, core, and being emphasized. It's important to note that Brats21 images are identical to Brats 20 images; the only difference is that the images are of different patients. Suppose our model can accurately predict the brain types and tumor stages of other patients with varying stages of tumor. This suggests that our model is reliable and effective for predicting these patients with various tumor stages. It's also crucial to note that sample 1 of the Brats21 dataset has a far larger tumor area than sample 2, even though the model correctly predicts both. The following outcomes were derived from the model's prediction of the class of brain tumors:
Fig. 21.
Brats21 Predicted image of sample 2.
Table 4 above shows that for many evaluation criteria, we were able to use the Brats21 dataset to predict samples with greater accuracy and dice coefficient values. This implies that the model is trustworthy and applicable to any dataset as long as it is pre-processed similarly to how we did in our study before being run through the model. To create arrays in the format that our model is trained with, we gathered samples from the Brats21 dataset and ran them through a generator. The arrays were then run through a trained model, and the results are shown in the table above. It appears that all evaluation metrics performed exceptionally well, demonstrating the robustness, efficacy, and accuracy of our DVIT-DSUNET model for brain tumor segmentation.
Table 4.
Model evaluation Performance for Brats21 Dataset.
| Evaluation Metric | Enhancing | EDEMA | Core | All classes |
|---|---|---|---|---|
| Test accuracy | 98.12 | 97.45 | 99.05 | 98.67 |
| Mean IOU | 0.1539 | 0.1487 | 0.1422 | 0.1563 |
| Dice Coefficient | 91.49 | 90.95 | 90.81 | 90.88 |
| Dice Loss | 0.0987 | 0.1012 | 0.0929 | 0.0912 |
| Precision | 98.15 | 98.98 | 99.09 | 98.91 |
| Sensitivity | 98.49 | 98.69 | 97.45 | 98.56 |
| Specificity | 99.19 | 98.98 | 98.17 | 98.77 |
| Blob Loss | 1.1298e-08 | 1.4535e-08 | 1.9854e-08 | 1.345e-08 |
The Test Accuracy is 98.67.
7. Discussion
Further, this work proposes a new approach to brain tumor segmentation using the Dual Vision Transformer-DSUNET model, which is new to the literature volume. Although the BRATS 2020 dataset is highly used in the literature, our technique is the first to employ dual vision transformers and a feature fusion approach to compare and contrast various MRI images (T1w, T2w, T1Gd, and FLAIR). This innovation greatly enhances the accuracy of the segmentation, where the Dice Coefficient scores obtained are 91. Seven percent for improved tumors, 92. 38 % for core tumors and 90 % for other tumors. 88 % for edema. The proposed model can be considered the first with automated brain tumor segmentation and serves as a solid groundwork for future studies.
To efficiently manage the dataset, we developed a unique MRI data generator to generate batches for training, validation, and testing. The model was better able to manage variations in the data that occur in the actual world thanks to the modifications used by the generator, such as Gaussian smoothing. Since the focused loss function effectively mitigates the issues brought on by a class imbalance in medical image segmentation, we adopted it as our preferred loss function during model training. With an incredible training accuracy of 99.93 %, the model performed wonderfully. Excellent accuracy, mean Intersection over Union (IoU), and great sensitivity and specificity are highlighted in the assessment criteria, which further support this achievement.
The Dual Vision Transformer-DSUNET's increased brain tumor segmentation performance has intriguing clinical implications. Tumor segmentation needs to be precise and efficient to enhance patient outcomes and create effective treatments. Our model offers the accuracy and memory necessary for medical imaging tasks, making it a useful tool for assisting radiologists in clinical practice.
In subsequent investigations, we want to strengthen the model's interpretability and address potential issues. More medical imaging datasets and research into additional preprocessing methods may help the model perform better across varied datasets. For the model to be verified and ultimately adopted, it must to be applied in a real-world clinical setting and evaluated by medical experts. This study proposes a new Dual Vision Transformer-DSUNET model, and its performance in segmenting brain tumors into their various components is demonstrated using the BRATS2020 dataset. The suggested model is a strong contender for accurate and efficient segmentation of brain tumors due to its exceptional accuracy and robustness. This research improves medical image processing and could significantly impact neuroimaging and clinical diagnosis.
The introduction of the Dual Vision Transformer-DSUNET architecture represents a significant advancement in brain tumor segmentation, particularly by implementing the dual attention mechanism. This approach intricately combines two distinct attentional techniques to enhance feature extraction and representation, which is crucial for accurate segmentation in medical imaging.
At the core of this architecture lies the dual attention block, an innovative element that integrates channel attention and spatial attention mechanisms. By learning channel-specific weights, channel attention focuses on identifying and emphasizing the most informative channels within the feature maps. This mechanism enables the model to prioritize channels contributing significantly to the task, effectively filtering out less relevant features. Doing so improves the model's ability to capture critical information from the data, which is essential for distinguishing between different tissue types or abnormalities in brain MRI scans.
Complementing channel attention is spatial attention, which directs the model's focus to the most relevant spatial regions within the feature maps. This spatially aware mechanism highlights essential areas crucial for accurate segmentation, ensuring that the model does not overlook key regions of interest. The synergy between these attentional techniques results in a more refined feature representation, as the model can simultaneously enhance the importance of specific channels and spatial regions.
The dual attention block's role extends beyond mere feature enhancement and serves as a crucial intermediate component within the network. Positioned between the encoder and decoder, this block acts as a bridge facilitating the transition of refined features through the network. By incorporating dual attention at this stage, the architecture ensures that the features passed to the decoder are both contextually relevant and rich in information, thereby improving the accuracy and effectiveness of the segmentation process.
The Dual Attention Fusion Block advances this approach by promoting collaboration between network streams. This block allows for the fusion of features from various nodes within the network, leveraging the strengths of each to produce a more comprehensive feature representation. By integrating dual attention mechanisms at this fusion stage, the model can effectively combine and refine features from multiple sources, ensuring that the most pertinent information is utilized for segmentation. This process enhances the model's ability to capture complex patterns and structures and improves its robustness in dealing with diverse and challenging imaging scenarios.
Overall, the dual attention mechanism employed in the Dual Vision Transformer-DSUNET architecture represents a significant step forward in medical image segmentation. By leveraging both channel and spatial attention techniques and incorporating a strategic fusion block, this approach enhances the model's capability to segment brain tumors accurately from MRI scans. Integrating these attentional mechanisms ensures that the model is better equipped to handle the intricacies of medical imaging data, ultimately leading to more precise and reliable segmentation results.
7.1. Discussion on dual attention
The proposed Dual Vision Transformer-DSUNET is a new architecture for brain tumor segmentation that has shown improvement with the added use of the dual attention mechanism. This approach very carefully intertwines two dissimilar attentional strategies to improve feature extraction and representation, which is highly important for segmentation in medical imaging applications.
One of the main components of this architecture is the dual attention block, which is a unique feature that combines channel attention and spatial attention blocks. Attention in channel pays attention to the locations of the channel within feature maps and assigns weights to the most informative channels. This mechanism allows the model to consider channels that make a big difference for the given task, thus excluding features that are not very important. In this way, it enhances the model's capacity for the extraction of crucial data in the differentiability of various tissue types or the identification of abnormalities in the brain MRI scans.
The other form of attention is spatial attention, which anchors the model's focus on the suitable spatial locations within the feature maps. This spatially aware mechanism points out significant areas for effective segmentation to avoid missing critical areas of interest. The combined effect of these attentional techniques helps to achieve a better feature representation, as the model can promote both channel and spatial attention at the same time.
The dual attention block also does not just act as a feature enhancement but is also a significant layer within the network. This block is placed between the encoder and decoder, and its’ primary role is to transfer the fine-grained features through the network. Including the dual attention in this step of the architecture enables the features to be passed to the decoder to be semantically related and packed with information density, thus helping the segmentation be more accurate and efficient.
The Dual Attention Fusion Block goes further than this by encouraging different network streams to work together. This block enables feature fusion from other nodes in the network, and this process compiles the strength of various features within the network. At this fusion stage, the employment of dual attention mechanisms will facilitate the combination and optimization of features from multiple sources so that only relevant information is exploited in segmentation. This process not only improves the descriptive and explanatory capability of the model in capturing multi-level and multi-scale patterns and structures within the image data but also increases the model's resistance to variations and variability of the imaging data.
In conclusion, the DA used in the architecture of Dual Vision Transformer-DSUNET can be regarded as a major advance in medical image segmentation. With the help of channel and spatial attention techniques and the addition of a fusion block, this model increases the accuracy of segmenting brain tumors using MRI scans. The integration of these attentional mechanisms allows the model to capture the complexities of medical images to enhance the accuracy and reliability of the segmentation maps.
7.2. Analysis of different parameters for the algorithms
When assessing tumor segmentation algorithms, we observe that our proposed model, the DVIT-DSUNET with feature fusion technique, outperforms the benchmarks across all types of tumors, as shown in Table 5. In particular, the proposed model yields dice accuracy of 0.908, 0.923, and 0.914 for Whole Tumor, Core Tumor, and Enhanced Tumor, respectively.
Table 5.
Different parameters for algorithms.
| Ref | Model | Whole Tumor | Core Tumor | Enhanced Tumor |
|---|---|---|---|---|
| Our Model | DVIT-DSUNET with feature fusion | 0.908 | 0.923 | 0.914 |
| [14] | Supervised Multi-Scale Attention Network | 0.902 | 0.869 | 0.806 |
| [17] | 3D-Znet | 0.906 | 0.845 | 0.859 |
| [21] | Swing BTS | 0.897 | 0.792 | 0.744 |
| [44] | Deep nuanced reasoning and Swin‐T | 0.920 | 0.872 | 0.913 |
| [45] | Dual Encoding- Decoding Method | 0.909 | 0.883 | 0.904 |
| [46] | Transformer and CNN combined | 0.908 | 0.853 | 0.857 |
| [47] | C-CNN with Distance-Wise Attention | 0.920 | 0.8726 | 0.9113 |
| [48] | CNN and Chimp Optimization Algorithm | 0.97 | – | – |
To summarise, whereas several models show impressive performance, our DVIT-DSUNET with feature fusion distinguishes itself by consistently achieving high DSC values in all tumor locations. This complete and balanced performance emphasizes the superiority of our model in efficiently managing the intricacies of tumor segmentation compared to the other studied methods.
Compared with the Supervised Multi-Scale Attention Network by Rehman et al. [14], we are getting higher performance for all categories. From the above analysis, Rehman et al. ’s model gets a score of 0.902 for the Whole Tumor, 0.869 for the Core Tumor, and 0.806 for the Enhanced Tumor, which accredits our enhanced ability to manage different and more complex characteristics of a tumor. Likewise, Ottom et al. ’s 3D-Znet [17], with the accuracy of 0.906, 0.845, and 0.859 in Core Tumor and Enhanced Tumor segmentation, are not as accurate as our model's segmentation capabilities due to our improved feature fusion techniques.
Compared with our approach, Jiang et al. ’s Swing BTS [21] has lower scores of 0.897, 0.792, and 0. 920 and 0. 913 in Whole and Enhanced Tumors, Segmentations in the Core Tumor area yields a score of 0. 872 which is far better than our model's 0.923. Li et al. ’s Dual Encoding-Decoding Method [45] gives comparatively better accuracy with 0.909, 0.883, and 0.904, though it is still less than our proposed model in the Core Tumor segmentation. Another work that performs less is Zhu et al. ’s [46] Transformer and CNN combined, which gets a dice accuracy of 0.908, 0.853, and 0.857. Implementing C-CNN with Distance-Wise Attention in Ref. [47] yields DSC scores of 0.920, 0.8726, and 0.9113. While the scores for the total tumor and improved tumor are competitive, the little difference in performance for the core tumor indicates that our model's feature fusion may provide a more sophisticated method for segmentation. Finally, Reference [48] utilizes Convolutional Neural Networks (CNN) in conjunction with the Chimp Optimization Algorithm, resulting in an exceptionally high DSC score of 0.97 for Whole Tumor. Nevertheless, there is a shortage of accessible data for core and expanded tumors. The lack of performance measurements for these crucial categories restricts the capacity to evaluate the model's efficacy thoroughly and complicates direct comparison. The outstanding Whole Tumor score indicates that although this model may perform exceptionally well in one aspect, it may not provide a holistic solution for overall tumor segmentation.
7.3. Comparative analysis
Seven research publications in this comparative analysis cover the segmentation of brain tumors using deep learning algorithms, as shown in Table 6. The papers emphasize increasing the precision and effectiveness of segmentation models for brain tumor detection.
Table 6.
Related studies for brain tumor segmentation.
| Reference | Approach | Dice Coefficient | Dataset |
|---|---|---|---|
| [14] | Supervised Multi-Scale Attention Network | 90 | BRATS dataset 2020 |
| [17] | 3D-Znet | 86 | BRATS dataset 2020 |
| [21] | Swing BTS | 82.85 | BRATS dataset 2020 |
| [23] | 2DU-Net | 89.9 | BRATS dataset 2020 |
| [28] | Residual Mix Transformer Fusion net. | 82.1 | BRATS dataset 2020 |
| [32] | Infinite Deformable Fusion Transformer Module (IDFTM) | 89.2 | BRATS dataset 2020 |
| [35] | PSwinBTS | 88.65 | BRATS dataset 2020 |
| [44] | Deep nuanced reasoning and Swin‐T | 89.1 | BRATS dataset 2020 |
| [45] | Dual Encoding- Decoding Method | 85.4 | BRATS dataset 2020 |
| [46] | Transformer and CNN combined | 90.63 | BRATS dataset 2019 |
| [47] | C-CNN with Distance-Wise Attention improves segmentation. | 91.13 | BRATS 2018 dataset |
| Our approach | DVIT-DSUNET with feature fusion | 91.25 | BRATS2020 |
Using a multi-scale attention network with selective deep supervision (SDS) mechanisms, the first study [14] introduces a technique known as SDS-MSANet. This method combines 2D and 3D inputs to retain sequential data. The design includes encoding units to extract global and local properties, attention units to weight coarse information, and a decoder block to scale up and learn tumor patterns. The suggested architecture improves segmentation performance, particularly for tumor regions that are core and enhancing. In this case, 90 % of the dice are thrown.
The 3D-Znet model, an enhanced deep neural network for segmenting 3D MR volumes, is presented in the second publication [17]. Dense connections are used in this model, based on the variational autoencoder-autodecoder Znet technique, to improve feature reuse. The architecture comprises input and output blocks, four encoders, and four decoders. Three-dimensional convolutional layers, batch normalization, and activation processes are all included in the encoder and decoder blocks. Results from the suggested 3D-Znet model are encouraging, with dice coefficient scores on par with cutting-edge methods. The dice result for this is 86 %.
The third study [21] proposes SwinBTS, a unique segmentation technique that incorporates an encoder-decoder structure, a transformer, and a convolutional neural network.
The network's encoder and decoder are a 3D Swin Transformer, and upsampling and downsampling are accomplished via convolutional processes. A better transformer module is included in the suggested model to extract specific features. According to extensive experimental results on numerous datasets, SwinBTS outperforms other 3D algorithms for brain tumor segmentation. The dice score is also 82.85 %.
The authors in reference used the same dataset using 2DU-Net (DoubleU-Net) [23]to produce a dice score of 89.9 %. The residual mix transformer fusion net is employed in Ref. [28], where it results in a dice score of 0.821. Also, using the Infinite Deformable Fusion Transformer Module (IDFTM) with PSwinBTS references [32,35]achieve dice scores of 89.2 % and 88.65 %, respectively.
Our method outperforms the others according to the Dice Coefficient measure when comparing our model to other approaches used to segment brain tumors. Ref [44] uses deep, subtle reasoning along with Swin Transformer (Swin-T), and the Dice score attained is 89.1. X-Net is introduced in Ref. [45], and the dual encoding-decoding method achieves a Dice score of 85.4. Also, Reference [46] integrates transformers and CNNs to merge deep semantics with edge information, thus obtaining a Dice score of 90.63 on the BRATS 2019 dataset. Additionally, reference [47] employs C-CNN with Distance-Wise Attention, and on the BRATS 2018 dataset, it has a dice coefficient of 91.13. Using distance-wise attention mechanisms improves the accuracy of segmentation. On the BRATS 2020 dataset, however, our DVIT-DSUNET outperforms this outcome with a Dice coefficient of 91.25, indicating better performance.
These references demonstrate additional strategies and how well they do segmentation. The research papers under discussion offer a variety of deep-learning algorithms for segmenting brain tumors. These models include cutting-edge methods like transformers, multi-scale attention networks, dense connections, and fusion modules. The experimental findings show enhanced segmentation capabilities for the core and augmenting regions of brain tumors. Compared to other cutting-edge methods, each model earns competitive dice scores and displays distinctive architectural design choices. These developments substantially impact medical image processing and may enhance the detection and management of brain cancers.
We devised and implemented the Dual Vision Transformer with feature fusion (DVIT-DSUNET) technology, greatly improving our investigation's brain tumor segmentation. This method outperforms several cutting-edge models, including the Selective Deeply Supervised Multi-Scale Attention Network, 3D-Znet, and SwinBTS, demonstrating its high technological viability and resilience. On the BRATS 2020 dataset, we achieved a remarkable Dice coefficient of 91.25 %, demonstrating the superior effectiveness of our strategy. Our Dice coefficient for enhanced tumors is 91.49 %, while that for core and all-class tumors is 90.81 % and 90.88 %, respectively, for model evaluation performance using the Brats dataset. Technically, our approach is viable due to the unique Deeply Supervised U-Net (DSUNET) and Dual Vision Transformer (DVIT) designs and practical feature fusion algorithms. To maximize the efficacy of transformers' self-attention mechanisms and U-Net's intrinsic ability to catch fine details in medical images, the DVIT-DSUNET technique utilizes the benefits of both architectures. This fusion enables our model to categorize data better using long-range linkages and multi-scale information.
The achievement of a higher dice coefficient compared to preceding state-of-the-art models further demonstrates the robustness of our method. By carefully integrating feature fusion, a U-Net-based architecture, and self-attention processes, our model demonstrates greater flexibility for complex oscillations in images of brain tumors. This adaptability produces more accurate and robust segmentation, even in challenging circumstances.
Our results advance the segmentation of brain tumors and show the potential of our approach to improve medical image analysis and help doctors make accurate diagnoses and treatment decisions.
7.4. Future directions
The results of our analysis suggest several exciting new lines of research and development. Numerous promising directions for expanding and strengthening our work include the ones listed below.
-
1.
Multi-Modal Fusion and Integration: To create a complete multi-modal brain tumor segmentation model, consider including other imaging modalities such as diffusion—and perfusion-weighted imaging (DWI) and perfusion-weighted imaging (PWI). Combining information from many modalities might give a more complete idea of the tumors' location.
-
2.
Uncertainty Quantification: The model's uncertainty estimate must be improved to improve clinical judgment. Use techniques like Monte Carlo dropout or Bayesian neural networks to determine how uncertain a forecast is. With trustworthy uncertainty estimates, clinicians can understand the model's level of assurance in its predictions.
-
3.
Explainability and Interpretability: Focus on making the model's predictions more understandable by using approaches to demonstrate and explain them. Understanding the model's decision-making process is necessary to win the trust and acceptance of the medical community.
-
4.
Transfer Learning and Domain Adaptation: Examine whether domain adaptation and transfer learning can improve the model's performance and make it more applicable to various datasets. Pretraining on an extensive dataset and fine-tuning on a smaller target dataset can improve efficiency and accuracy.
-
5.
Clinical Validation and Deployment: Collaborate with healthcare specialists to properly evaluate the model in clinical situations. Gather information from the real world and conduct thorough testing in healthcare environments. Address any problems and recommendations to improve the model for use in a clinical environment.
Fig. 22 depicts the sequential stages involved in the prospective advancements in identifying brain tumors.
-
6.
Interactive Interface for Radiologists: Create a user-friendly interface to allow radiologists to engage with and provide feedback on the model's predictions. This feedback loop can help with iterative model improvement and ensure that it complies with physicians' practical needs.
-
7
Integration with Clinical Decision Support Systems: Develop clinical decision support systems (CDSS) that leverage the model to assist radiologists in their daily activities. The model's predictions can be an additional data source, allowing for more accurate and potent diagnoses.
-
8.
Incorporate 3D Architectures: To fully utilize the volumetric character of medical imaging data, investigate and design 3D structures. Particularly in the case of brain tumor segmentation, 3D models may be able to represent spatial relationships more precisely.
-
9.
Robustness to Noisy and Incomplete Data: You may improve the model's capacity to handle noisy or missing input by experimenting with data imputation techniques and adversarial training. This will ensure the model's trustworthiness in scenarios when the data quality may change.
-
10.
Collaborative Research and Benchmarking: To create standards for assessment measures and benchmarks and promote cooperation with other research teams. A comparison study with other cutting-edge models will provide a deeper understanding of our strategy's benefits and drawbacks and also provide a path for future advancements.
Fig. 22.
Future directions in brain tumor detection.
By following these potential future directions, we can advance the understanding of brain tumor segmentation and produce more precise, dependable, and therapeutically beneficial tools to assist medical professionals in identifying and treating brain tumors.
7.5. Core contributions
Our utilization of the BRATS2020 dataset has resulted in a significant advancement in brain tumor segmentation. As depicted in Fig. 23, our research's primary contributions can be succinctly summarized as follows.
-
1.
Dual Vision Transformer Architecture: We developed a novel architecture, the Dual Vision Transformer (DVIT), to segregate brain malignancies. This design combines the Transformer model, well-known for its efficiency in natural language processing, with convolutional neural networks (CNNs). The DVIT design is more effective at gathering regional and international components of medical imaging data.
-
2.
DSUNET Model with Feature Fusion: We developed a sophisticated DSUNET (Dual Stream U-Net) with feature fusion for the segmentation of brain tumors. Employing dual attention techniques in the DSUNET variation of the U-Net architecture enhances the model's ability to focus on important areas. We also implemented feature fusion to utilize complementary data from several network sources.
-
3.
Incorporation of Dual Attention Mechanisms: We added channel attention and spatial attention to our model as dual attention processes. By adaptively stressing essential components, these techniques enable the model to minimize noise while enhancing its ability to distinguish effectively.
-
4.
Feature Fusion for Enhanced Representation: Feature fusion combines features from multiple network parts. This fusion technique facilitates the representation of features and generates feature maps that are more informative and discriminative by combining data from many channels.
-
5.
Utilization of the BRATS2020 Dataset: We used the BRATS2020 dataset, a frequently used benchmark dataset, to categorize brain tumors. Using this dataset allowed us to perform thorough testing, validate our models, and ensure their robustness and generalizability.
-
6.
Implementation of Focal Loss: We used the targeted loss function during model training, focusing on hard-to-classify samples, to lessen the consequences of class imbalance. The focus loss function helped achieve high training accuracy and properly handle the data imbalance inherent in medical imaging datasets.
-
7.
Efficient Data Pre-Processing Strategies: We used the targeted loss function during model training, focusing on hard-to-classify samples, to lessen the consequences of class imbalance. The focus loss function helped us properly handle the data imbalance inherent in medical imaging datasets, achieving high training accuracy.
-
8.
Implementation of Focal Loss: We used the targeted loss function during model training, focusing on hard-to-classify samples, to lessen the consequences of class imbalance. The focus loss function helped achieve high training accuracy by properly handling the data imbalance inherent in medical imaging datasets.
-
9.
Efficient Data Pre-Processing Strategies: We prepared the input data for the model using several effective data preparation methods, including intensity normalization, scaling, and one-hot encoding. These preprocessing steps ensured consistency and enhanced the model's ability to gather valuable data.
-
10.
Comprehensive Model Evaluation: Accuracy, mean, and loss We thoroughly tested our models with the intersection over Union (IoU), dice coefficient, accuracy, sensitivity, and specificity metrics. Thanks to this in-depth examination, we understood the model's performance in several areas.
-
11.
Detailed Experimentation and Analysis: We thoroughly examined a variety of model topologies, hyperparameters, and preprocessing methods. We gave the findings significant thought, providing enlightening information on the advantages and disadvantages of our strategy and directing continued development.
-
12.
Introduction of Novel Loss: Blob loss directly addresses the issue of disconnected or disjointed segmentation results. Medical imaging requires precise segmentation because disconnected parts typically result in inaccurate representations of anatomical features. Blob loss penalizes such fragmented segments, encouraging the algorithm to produce more continuous and coherent segmentations. Blob loss improves the segmentation model's resilience by limiting the formation of isolated blobs.
Fig. 23.
Core contributions of our study.
These significant contributions demonstrate improved model design, attention processes, data preparation, and comprehensive assessment techniques. They also demonstrate the significance of our work in brain tumor segmentation and establish a strong foundation in this essential field of healthcare research and innovation.
8. Conclusion
The present work proposes a new method for segmenting brain tumors using the Dual Vision Transformer-DSUNet with feature fusion. This method also improves the accuracy and reliability of medical image analysis. The investigation showcases that the integration of dual attention mechanisms with feature fusion strategies significantly enhances the model's ability to learn complicated and long-distance connections, pay attention to relevant features, and achieve better segmentation results.
The proposed Dual Vision Transformer-DSUNet model has shown high effectiveness in training for accurate segmentation of brain tumors with a training accuracy of 99. 93 %. The higher precision they demonstrate proves the model's high ability to distinguish between the areas with tumors and those without them. Further, the results like Dice Coefficient, IoU, Sensitivity, Specificity, Precision, Recall, F1 Score, etc., also justify the higher accuracy and reliability of the proposed model in real-world applications. Further, the advantages of the proposed Dual Vision Transformer-DSUNet are remarkable in the use of dual attention mechanisms that help to address the problem of the imbalance in the distribution of classes and focus on the most relevant features. This is a precious capacity since the precision of the measurements is critical in medical imaging. From this perspective, by bringing the ability of class distribution into equilibrium during training, the actual equilibrium and efficiency of the model are improved to offer more accurate results in segmentation.
Moreover, the combination of the Dual Vision Transformer with the use of the U-Net model approach takes advantage of each approach. The ability to gather such detailed information in the U-Net and the ability of the Transformer to understand global, complex relationships between such issues also help create a strong tool for segmenting brain tumors. Not only does this approach enhance the accuracy of the segmentation methodology, but also it can be adapted to the various challenges faced in medical imaging. Likewise, the results of the present study stress the vast potentiality of the Dual Vision Transformer (DSUNet) in clinical scenarios. Due to the ability of this model to offer a better and more reliable way of segmenting brain tumor regions than the previous models, it has the potential to improve diagnostic outcomes and the planning of treatment procedures. This study also stresses its applicability in clinical practice to help practitioners make better decisions and enhance patient outcomes.
Moreover, this study is helpful for future developments in the medical imaging field that will be advanced. The effectiveness of deep learning architecture and employing advanced techniques can be further studied and enhanced based on the prior research findings. The following are some recommendations for future studies: First, increase the model's computational speed. Second, expand the model to different types of medical images and different types of diseases.
Lastly, the proposed Dual Vision Transformer-DSUNet with feature fusion is another remarkable technological advancement in brain tumor segmentation. Combining advanced deep learning techniques with new attention mechanisms is a solid ground for improving the interpretation of medical images. By continued refinement, this approach aims to provide major enhancements to the field, enhancing patient care and treatment outcomes by increasing the accuracy of diagnosis and the efficacy of the strategic therapeutic planning process.
8.1. Future research
Last but not least, more excellent investigation into the Dual Vision Transformer-DSUNet with feature fusion for brain tumor segmentation may result in significant advancements in the medical imaging area. The fusion of multi-modal imaging data is one potential future research direction. The segmentation of brain tumors using a single-modality MRI dataset was the main goal of the current investigation. Supplementary imaging techniques like CT scans or PET scans might be used to understand the tumor and its boundaries better. Researchers may obtain more precise tumor segmentation by utilizing the information in multi-modal data and the Dual Vision Transformer-DSUNet's capacity to record long-range associations. Further research into the model's performance on more extensive, varied datasets is also necessary.
Although the results of the current study were encouraging, the approach's robustness and generalizability would be better established by testing it on a more extensive and varied dataset. Testing the model on diverse brain tumor types while considering size, shape, and location changes would be required. Additionally, analyzing the model's performance on datasets from various clinics and imaging systems would offer insightful information about its suitability in clinical settings. Future studies should also focus on investigating other attention mechanisms and fusion strategies. Finding critical characteristics for segmenting the tumor into different parts was accomplished using the two-level attention technique in this study. Other attention mechanisms, such as multi-head or self-attention, are nevertheless available and may improve segmentation accuracy further.
Similarly, investigating various feature fusion methods, such as densely connected networks or skip connections, could enhance the model's performance even further. In conclusion, there is a lot of potential for the Dual Vision Transformer-DSUNet with feature fusion for brain tumor segmentation to transform medical imaging. By using multi-modal data, evaluating performance on more extensive and more diverse datasets, and investigating various attention mechanisms and fusion strategies, researchers might advance the field of brain tumor segmentation. Better brain tumor diagnosis and patient treatment planning will eventually result from this.
Ethical approval
Not applicable.
Funding
Researchers Supporting Project number (RSPD2024R1107), King Saud University, Riyadh, Saudi Arabia.
Data availability statement
The dataset is available on reasonable request.
CRediT authorship contribution statement
Mohammed Zakariah: Writing – original draft, Supervision, Software, Formal analysis, Data curation, Conceptualization. Muna Al-Razgan: Validation, Methodology, Investigation, Funding acquisition. Taha Alfakih: Writing – review & editing, Visualization, Validation.
Declaration of competing interest
The authors declare there is no conflict of interest.
Acknowledgments
The authors would like to thank Researchers Supporting Project number (RSPD2024R1107), King Saud University, Riyadh, Saudi Arabia.
Contributor Information
Mohammed Zakariah, Email: mzakariah@ksu.edu.sa.
Muna Al-Razgan, Email: malrazgan@ksu.edu.sa.
Taha Alfakih, Email: talfakih@ksu.edu.sa.
References
- 1.Srikanth M.V., Prasad V.V.K.D.V., Satya Prasad K. Brain tumor detection through modified optimization algorithm by region-based image fusion. ECTI Transactions on Computer and Information Technology (ECTI-CIT) Mar. 2023;17(1):117–127. doi: 10.37936/ecti-cit.2023171.249604. [DOI] [Google Scholar]
- 2.Preethi S., Aishwarya P. An efficient wavelet-based image fusion for brain tumor detection and segmentation over PET and MRI image. Multimed Tools Appl. 2021;80(10):14789–14806. doi: 10.1007/s11042-021-10538-3. [DOI] [Google Scholar]
- 3.Fang L., Wang X. Brain tumor segmentation based on the dual-path network of multi-modal MRI images. Pattern Recognit. Apr. 2022;124 doi: 10.1016/j.patcog.2021.108434. [DOI] [Google Scholar]
- 4.Hamdaoui F., Sakly A. Automatic diagnostic system for segmentation of 3D/2D brain MRI images based on a hardware architecture. Microprocess. Microsyst. Apr. 2023;98 doi: 10.1016/j.micpro.2023.104814. [DOI] [Google Scholar]
- 5.Shen T., Xu H. Medical image segmentation based on transformer and HarDNet structures. IEEE Access. 2023;11:16621–16630. doi: 10.1109/ACCESS.2023.3244197. [DOI] [Google Scholar]
- 6.Wang Z., Zou Y., Liu P.X. Hybrid dilation and attention residual U-Net for medical image segmentation. Comput. Biol. Med. Jul. 2021;134 doi: 10.1016/j.compbiomed.2021.104449. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Mu F., Shi Y., Cheng J., Li C., Chen X. Brain tumor segmentation in multimodal MRI via pixel-level and feature-level image fusion. Front. Neurosci. 2022;16 doi: 10.3389/fnins.2022.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y., Wang J. 2022 IEEE 35th International Symposium on Computer-Based Medical Systems (CBMS) IEEE; 2022. TSEUnet: a 3D neural network with fused Transformer and SE-Attention for brain tumor segmentation; pp. 131–136. [DOI] [Google Scholar]
- 9.Samosir R.S., Abdurachman E., Gaol F.L., Sabarguna B.S. Brain tumor segmentation using double density dual tree complex wavelet transform combined with convolutional neural network and genetic algorithm. IAES Int. J. Artif. Intell. 2022;11(4):1373. doi: 10.11591/ijai.v11.i4.pp1373-1383. [DOI] [Google Scholar]
- 10.R J., D S. 2020. Segmentation Techniques for Brain Tumor from MRI – A Survey. [DOI] [Google Scholar]
- 11.Cui H., Yuwen C., Jiang L., Xia Y., Zhang Y. Multiscale attention guided U-Net architecture for cardiac segmentation in short-axis MRI images. Comput Methods Programs Biomed. Jul. 2021;206 doi: 10.1016/j.cmpb.2021.106142. [DOI] [PubMed] [Google Scholar]
- 12.Asiri A.A., et al. Advancing brain tumor classification through fine-tuned vision transformers: a comparative study of pre-trained models. Sensors. Sep. 2023;23(18):7913. doi: 10.3390/s23187913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vijithananda S.M., et al. Feature extraction from MRI ADC images for brain tumor classification using machine learning techniques. Biomed. Eng. Online. Dec. 2022;21(1):52. doi: 10.1186/s12938-022-01022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rehman A., Usman M., Shahid A., Latif S., Qadir J. Selective deeply supervised multi-scale attention network for brain tumor segmentation. Sensors. 2023;23(4):2346. doi: 10.3390/s23042346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X., Yang L. Proceedings of the 1st ACM Workshop on Mobile and Wireless Sensing for Smart Healthcare. ACM; New York, NY, USA: 2022. Brain tumor segmentation based on CBAM-TransUNet; pp. 33–38. [DOI] [Google Scholar]
- 16.Anaya-Isaza A., Mera-Jiménez L., Fernandez-Quilez A. CrossTransUnet: a new computationally inexpensive tumor segmentation model for brain MRI. IEEE Access. 2023;11:27066–27085. doi: 10.1109/ACCESS.2023.3257767. [DOI] [Google Scholar]
- 17.Ottom M.A., Abdul Rahman H., Alazzam I.M., Dinov I.D. Multimodal stereotactic brain tumor segmentation using 3D-znet. Bioengineering. 2023;10(5):581. doi: 10.3390/bioengineering10050581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajasree R., Columbus C.C., Shilaja C. Multiscale-based multimodal image classification of brain tumor using deep learning method. Neural Comput. Appl. Jun. 2021;33(11):5543–5553. doi: 10.1007/s00521-020-05332-5. [DOI] [Google Scholar]
- 19.Amin J., Sharif M., Haldorai A., Yasmin M., Nayak R.S. Brain tumor detection and classification using machine learning: a comprehensive survey. Complex & Intelligent Systems. Aug. 2022;8(4):3161–3183. doi: 10.1007/s40747-021-00563-y. [DOI] [Google Scholar]
- 20.Muezzinoglu T., et al. PatchResNet: multiple patch division–based deep feature fusion framework for brain tumor classification using MRI images. J. Digit. Imag. Feb. 2023;36(3):973–987. doi: 10.1007/s10278-023-00789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y., Zhang Y., Lin X., Dong J., Cheng T., Liang J. SwinBTS: a method for 3D multimodal brain tumor segmentation using Swin transformer. Brain Sci. 2022;12(6):797. doi: 10.3390/brainsci12060797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandya P., Oguine O.C., Bhargava H., Zade S. Enhanced 3D brain tumor segmentation using assortedprecision training. BOHR International Journal of Internet of things, Artificial Intelligence and Machine Learning. 2022;1(1):65–69. doi: 10.54646/bijiam.2022.10. [DOI] [Google Scholar]
- 23.L D.P., Ponraj D.N. 2023 4th International Conference on Signal Processing and Communication (ICSPC) IEEE; 2023. Modified 2D-UNet for brain tumor segmentation; pp. 229–233. [DOI] [Google Scholar]
- 24.Zhao J., Dang M., Chen Z., Wan L. DSU-Net: distraction-Sensitive U-Net for 3D lung tumor segmentation. Eng. Appl. Artif. Intell. Mar. 2022;109 doi: 10.1016/j.engappai.2021.104649. [DOI] [Google Scholar]
- 25.Borlea I.-D., Precup R.-E., Borlea A.-B., Iercan D. A unified form of fuzzy C-means and K-means algorithms and its partitional implementation. Knowl. Base Syst. Feb. 2021;214 doi: 10.1016/j.knosys.2020.106731. [DOI] [Google Scholar]
- 26.Al-Zoghby A.M., Al-Awadly E.M.K., Moawad A., Yehia N., Ebada A.I. Dual deep CNN for tumor brain classification. Diagnostics. Jun. 2023;13(12):2050. doi: 10.3390/diagnostics13122050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfister S.M., et al. A summary of the inaugural WHO classification of pediatric tumors: transitioning from the optical into the molecular era. Cancer Discov. Feb. 2022;12(2):331–355. doi: 10.1158/2159-8290.CD-21-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gai D., Zhang J., Xiao Y., Min W., Zhong Y., Zhong Y. RMTF-net: residual mix transformer fusion net for 2D brain tumor segmentation. Brain Sci. 2022;12(9):1145. doi: 10.3390/brainsci12091145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shafi A.S.M., Rahman Md B., Anwar T., Halder R.S., Kays H.M.E. Classification of brain tumors and auto-immune disease using ensemble learning. Inform. Med. Unlocked. 2021;24 doi: 10.1016/j.imu.2021.100608. [DOI] [Google Scholar]
- 30.Choudhury C.L., Mahanty C., Kumar R., Mishra B.K. 2020 International Conference on Computer Science, Engineering and Applications (ICCSEA) IEEE; Mar. 2020. Brain tumor detection and classification using convolutional neural network and deep neural network; pp. 1–4. [DOI] [Google Scholar]
- 31.Badža M.M., Barjaktarović M.Č. Classification of brain tumors from MRI images using a convolutional neural network. Appl. Sci. Mar. 2020;10(6):1999. doi: 10.3390/app10061999. [DOI] [Google Scholar]
- 32.Nian Z.R., et al. ArXiv (Cornell University); 2023. 3D Brainformer: 3D Fusion Transformer for Brain Tumor Segmentation. [Google Scholar]
- 33.Lin J., et al. CKD-TransBTS: clinical knowledge-driven hybrid transformer with modality-correlated cross-attention for brain tumor segmentation. IEEE Trans Med Imaging. 2023;42(8):2451–2461. doi: 10.1109/TMI.2023.3250474. [DOI] [PubMed] [Google Scholar]
- 34.Arora G.K., Taj Mahaboob S., Adilakshmi S., Rani S.K., Kasthuri A., Mamodiya U. 2023 1st International Conference on Innovations in High Speed Communication and Signal Processing (IHCSP) IEEE; 2023. Establishment of an effective brain tumor classification system through image transformations and optimization techniques; pp. 12–16. [DOI] [Google Scholar]
- 35.Zongren L., Silamu W., Shurui F., Guanghui Y. Focal cross transformer: multi-view brain tumor segmentation model based on cross window and focal self-attention. Front. Neurosci. 2023;17 doi: 10.3389/fnins.2023.1192867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu A., Razmjooy N. Brain tumor diagnosis based on metaheuristics and deep learning. Int J Imaging Syst Technol. 2021;31(2):657–669. doi: 10.1002/ima.22495. [DOI] [Google Scholar]
- 37.Amin J., Sharif M., Yasmin M., Saba T., Anjum M.A., Fernandes S.L. A new approach for brain tumor segmentation and classification based on score level fusion using transfer learning. J. Med. Syst. 2019;43(11):326. doi: 10.1007/s10916-019-1453-8. [DOI] [PubMed] [Google Scholar]
- 38.El‐Hag N.A., et al. Utilization of image interpolation and fusion in brain tumor segmentation. Int J Numer Method Biomed Eng. 2021;37(8) doi: 10.1002/cnm.3449. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Z., Sun M., Qi G., Li Y., Gao X., Liu Y. Sparse Dynamic Volume TransUNet with multi-level edge fusion for brain tumor segmentation. Comput. Biol. Med. 2024;172:108284. doi: 10.1016/j.compbiomed.2024.108284. 108284. [DOI] [PubMed] [Google Scholar]
- 40.Yue G., Zhuo G., Zhou T., Liu W., Wang T., Jiang Q. Adaptive cross-feature fusion network with inconsistency guidance for multi-modal brain tumor segmentation. IEEE Journal of Biomedical and Health Informatics. 2023:1–11. doi: 10.1109/jbhi.2023.3347556. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y., Mu F., Shi Y., Chen X. SF-net: a multi-task model for brain tumor segmentation in multimodal MRI via image fusion. IEEE Signal Process. Lett. 2022;29:1799–1803. doi: 10.1109/lsp.2022.3198594. [DOI] [Google Scholar]
- 42.Berlin Shaheema S., Suganya Devi K., Naresh Babu Muppalaneni. Explainability based Panoptic brain tumor segmentation using a hybrid PA-NET with GCNN-ResNet50. Biomed. Signal Process Control. 2024;94:106334. doi: 10.1016/j.bspc.2024.106334. 106334. [DOI] [Google Scholar]
- 43.Yue G., Wu S., Du J., Zhou T., Jiang B., Wang T. Adaptive context aggregation network with prediction-aware decoding for multi-modal brain tumor segmentation. IEEE Trans. Instrum. Meas. 2024:1. doi: 10.1109/tim.2024.3417601. 1. [DOI] [Google Scholar]
- 44.Xu Y., Yu K., Qi G., Gong Y., Qu X., Yin L., Yang P. Brain tumour segmentation framework with deep nuanced reasoning and Swin‐T. IET Image Process. 2024 doi: 10.1049/ipr2.13048. [DOI] [Google Scholar]
- 45.Li Y., Wang Z., Li Y., Zhu Z., Qi G., Liu Y. X-Net: a dual encoding–decoding method in medical image segmentation. Vis. Comput. 2021;39(6):2223–2233. doi: 10.1007/s00371-021-02328-7. [DOI] [Google Scholar]
- 46.Zhu Z., He X., Yu Hen Hu, Li Y., Cong B., Liu Y. Brain tumor segmentation based on the fusion of deep semantics and edge information in multimodal MRI. 2023;91:376. doi: 10.1016/j.inffus.2022.10.022. 387. [DOI] [Google Scholar]
- 47.Ranjbarzadeh R., Bagherian Kasgari A., Jafarzadeh Ghoushchi S., Anari S., Naseri M., Bendechache M. Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-90428-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ranjbarzadeh R., Zarbakhsh P., Caputo A., Tirkolaee E.B., Bendechache M. Brain tumor segmentation based on optimized convolutional neural network and improved chimp optimization algorithm. Comput. Biol. Med. 2024;168 doi: 10.1016/j.compbiomed.2023.107723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset is available on reasonable request.