FIGURE 4.
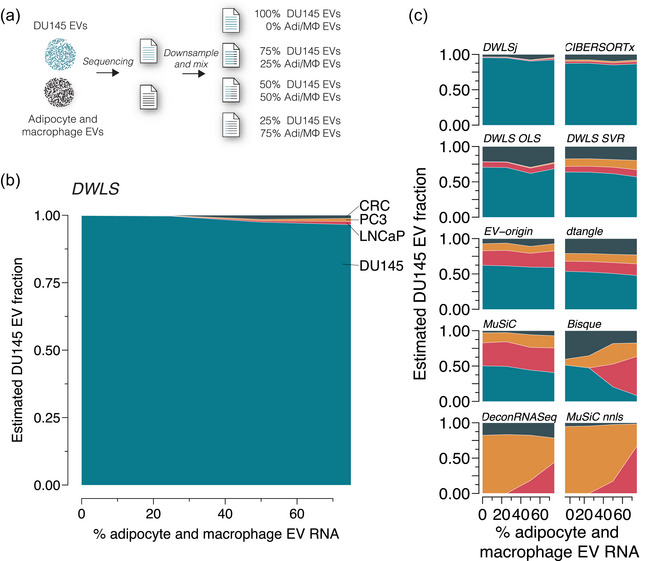
Effect of missing cell types in the signature matrix. (a) In silico mixed EV samples were created by downsampling and mixing RNA reads from DU145 and physically mixed adipocyte and macrophage EV samples (GSE94155) in ratios of 100:0, 75:25, 50:50, 25:75 (n = 3 for each ratio). The samples were deconvoluted using the transcript signatures for DU145, LNCaP, PC3 and CRC cells. (b) The adipocyte and macrophage EV RNA did not significantly affect the cell type‐specific EV fraction estimated by DWLS. (c) Similarly, the cell‐specific EV estimated of the mixed samples were not significantly affected by adipocyte and macrophage EV RNA for DWLSj, CIBERSORTx, DWLS OLS, DWLS SVR, EV‐origin, dtangle and MuSiC deconvolution methods; however, the presence of 50% or more adipocyte and macrophage EV RNA changed cell type‐specific EV estimates for Bisque, DeconRNASeq and MuSiC nnls deconvolution methods.
