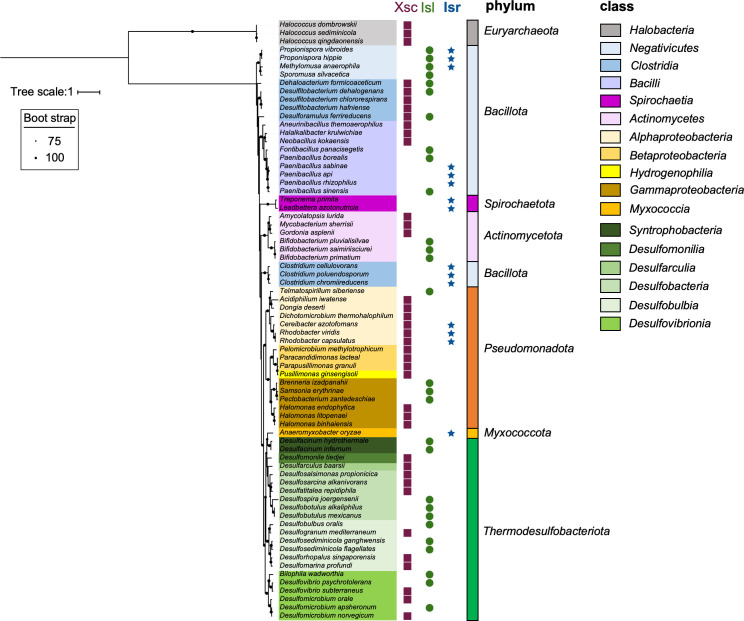
Fig 8.
Distribution of the three oxygen-independent isethionate metabolic enzymes (Isr, Isl, and Xsc) in prokaryotes. Distribution of the three enzymes Xsc (purple squares), IslA (green circles), and IsrD (blue stars), which are oxygen-independent enzymes for isethionate metabolism, are plotted on the phylogenetic tree of 16 s rRNA. The best fit model is TIM3 +F + I + G4.