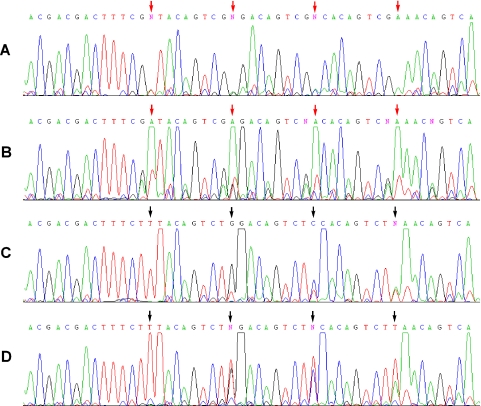
Figure 4.
Dye terminator sequencing of two ODNs used to examine dMeisoC and disoG in all possible natural nearest-neighbor contexts (Supplementary Figures S6 and S7 contain additional sequences). An ODN containing four dMeisoC positions (red arrows) was sequenced in a BigDye 3.1 reaction in the presence of (A) 300 μM disoGTP and 0 μM dMeisoCTP or (B) 0 μM disoGTP and 0 μM dMeisoCTP. The template sequence in the region shown is 3′-TGCTGCTGAAAGCiCATGTCAGCiCCTGTCAGCiCGTGTCAGCiCTTTGTCAG-5′. Very large ddA terminator signals were always observed opposite dMeisoC in the absence of disoGTP, perhaps indicating a transition state base pairing geometry different from complementary pairings. An ODN containing four disoG positions (black arrows) was sequenced in a BigDye 3.1 sequencing reaction in the presence of (C) 100 μM dMeisoCTP and 0 μM disoGTP or (D) 0 μM dMeisoCTP and 0 μM disoGTP. The template sequence in the region shown is 3′-TGCTGCTGAAAGAiG ATGTCAGAiGCTGTCAGAiGGTGTCAGAiGTTGTCAG-5′. Terminator ddT was always incorporated opposite disoG in the absence of dMeisoCTP. Extremely large terminator signals were observed at the position following disoG for all conditions. Additionally, a noticeable signal that may correspond to the polymerase skipping over a fraction of template disoG positions was visible opposite disoG. Little, if any, signal attenuation caused by unwanted strand termination was visible upon encountering isolated dMeisoC or disoG positions, either in the presence or absence of complements disoGTP and dMeisoCTP. This contrasts with Figure 3C, in which several proximate non-natural template positions caused premature termination of sequencing reactions lacking disoGTP and dMeisoCTP.