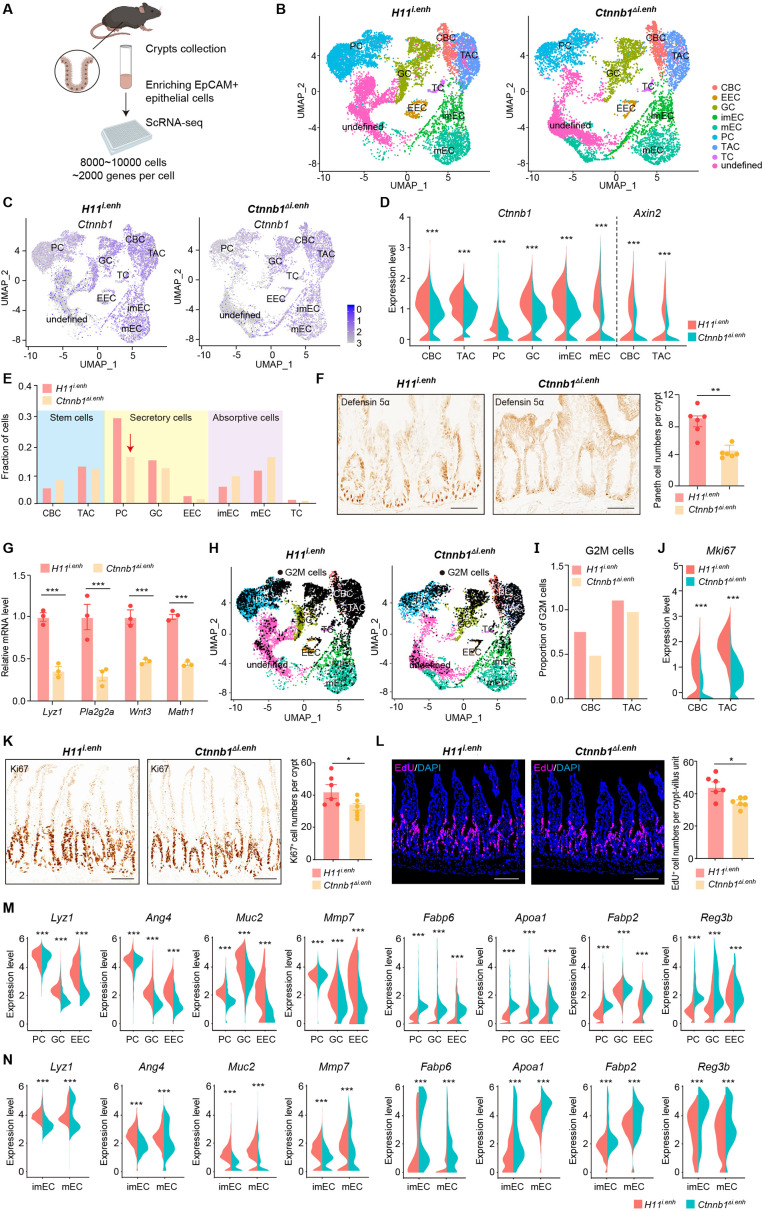
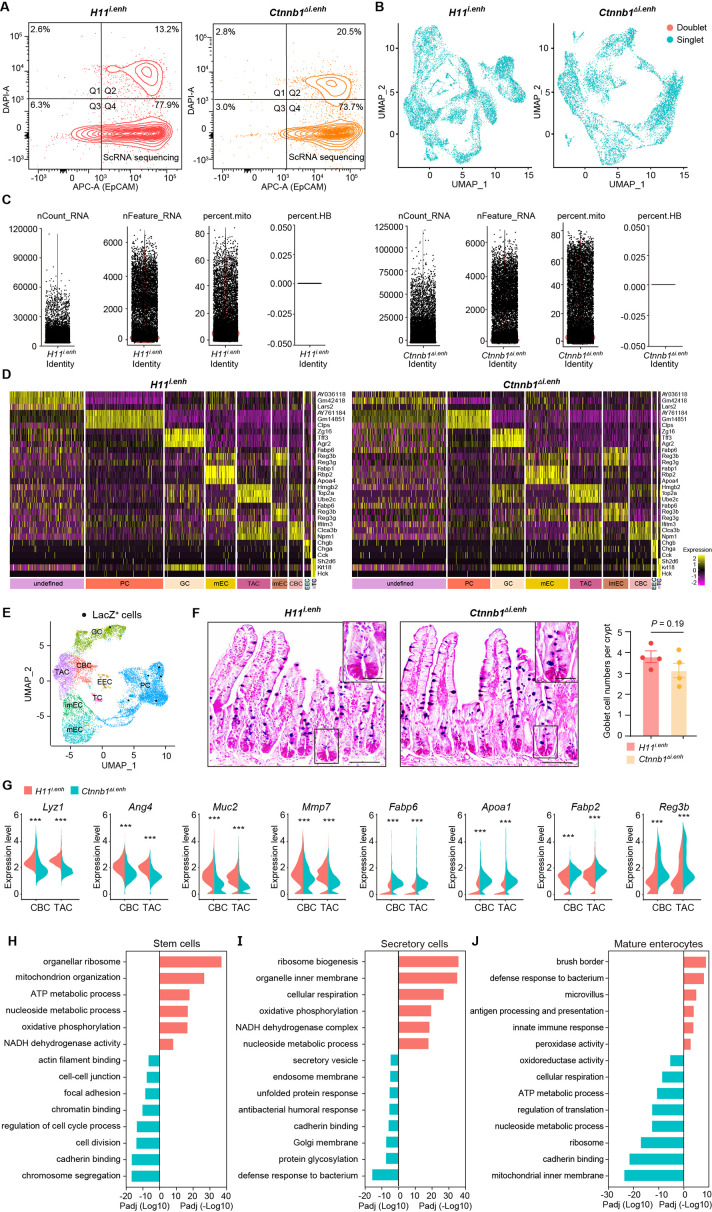
Figure 2. ieCtnnb1 knockout altered cellular composition and expression profiles of small intestinal crypts.
(A) Schematic illustration of single-cell sequencing. Crypts were extracted from small intestines followed by fluorescence activated cell sorting (FACS) to enrich EpCAM+ DAPI- epithelial cells. Cells of two 10-week-old female mice for each genotype were pooled together to perform single-cell transcriptome analyses. (B) Uniform Manifold Approximation and Projection (UMAP) were used to visualize the clustering of 11,824 single cells from H11i.enh mice and 8,094 single cells from Ctnnb1Δi.enh mice. Cell types were assigned according to expressions of marker genes. CBC, crypt base columnar cell; TAC, transit-amplifying cell; EEC, enteroendocrine cell; imEC, immature enterocytes; mEC, mature enterocytes; GC, goblet cell; PC, Paneth cell; TC, tuft cell. (C) Expression and distribution of Ctnnb1 in small intestinal crypt cells of H11i.enh and Ctnnb1Δi.enh mice. (D) Violin plots showing the expression of Ctnnb1 in CBC, TAC, PC, GC, imEC, mEC; and the expression of Axin2 in CBC and TAC, of H11i.enh and Ctnnb1Δi.enh mice. (E) Comparison of the proportion of indicated small intestinal crypt cell types in H11i.enh and Ctnnb1Δi.enh mice. (F) Immunohistochemistry (left and middle) and quantification (right) of PCs in small intestines of H11i.enh (n=6) and Ctnnb1Δi.enh (n=6) mice. (G) Quantitative reverse transcription PCR (RT-qPCR) showing relative mRNA levels of PC marker genes (Lyz1, Pla2g2a, Wnt3, Math1) in small intestinal crypts of H11i.enh (n=3) and Ctnnb1Δi.enh (n=3) mice. (H) Distribution of G2M cells in H11i.enh and Ctnnb1Δi.enh small intestinal crypts, based on the expression of cell cycle marker gene Mki67. (I) Comparison of the proportion of G2M cells in CBC and TAC of H11i.enh and Ctnnb1Δi.enh small intestinal crypts. (J) Violin plots showing the expression of Mki67 in CBC and TAC of H11i.enh and Ctnnb1Δi.enh small intestinal crypts. (K) Immunohistochemistry (left and middle) and quantification (right) of Ki67+ cells in small intestines of H11i.enh (n=6) and Ctnnb1Δi.enh (n=6) mice. (L) Immunofluorescence (left and middle) and quantification (right) of EdU+ cells (red) in small intestines of H11i.enh (n=6) and Ctnnb1Δi.enh (n=6) mice after 4 hr EdU injection. Nuclei were labeled with DAPI (blue). (M–N) Violin plots showing expressions of marker genes for secretory cells (Lyz1, Ang4, Muc2, Mmp7) and absorptive cells (Fabp6, Apoa1, Fabp2, Reg3b) in secretory(M) and absorptive lineages (N) of H11i.enh and Ctnnb1Δi.enh small intestinal crypts. Scale bars, 50 μm (F, K, and L). Quantification data are shown as means ± SEM, statistical significance was determined using an unpaired two-tailed Student’s t-test (D, F, J, K, L, M, and N). Quantification data are shown as means ± SD, statistical significance was determined using Multiple t-tests – one per row (G). *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. ns, not significant.