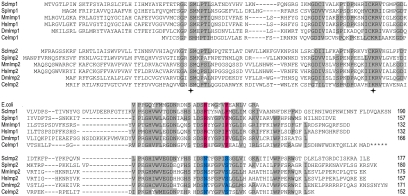
Figure 5.
Multiple sequence alignment of the Imp family of proteases. Iterative BLAST analysis was undertaken with the Imp2 sequence from S. cerevisiae. The Imp1 and Imp2 sequences were aligned with ClustalW, and a representative selection is shown (Sc, S. cerevisiae; Sp, Schizosaccharomyces pombe, Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, M. musculus). Asterisks designate a possible C-terminal extension to CeImp1. Amino acid residues conserved across at least six species are highlighted, and the total number of residues shown. Imp1-specific residues (RX5P) are colored pink and Imp2-specific residues (NX5S) are colored blue. Catalytic residues in Imp1 and Imp2 (analogous to Ser90 and K145 of E. coli leader peptidase) are designated with stars.