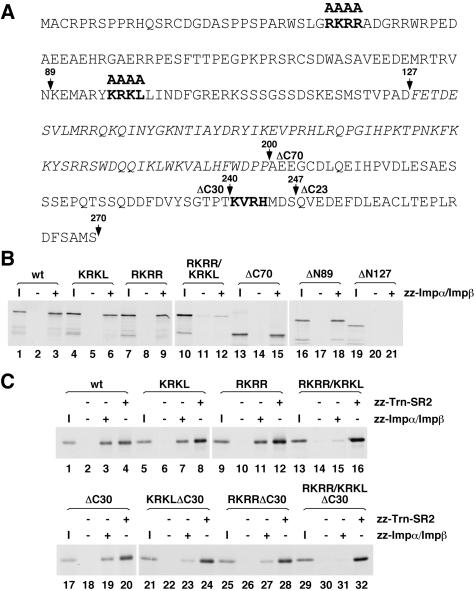
Figure 4.
Mapping the sequences in SLBP required for Impα/Impβ binding. (A) The amino acid sequence of human SLBP is shown with the RNA binding domain in italics. The predicted Impα binding sites that were mutated to alanines are in bold. The arrows above the sequence mark the amino acids at which the protein was truncated either from the N-terminal end (ΔN89 and ΔN127) or the C-terminal end (ΔC70 and ΔC30). (B) The different in vitro-translated SLBP truncation or site-specific mutants labeled with [35S]methionine were incubated without import receptor (lanes 2, 5, 8, 11, 14, 17, and 20) or with zz-Impα/Impβ (lanes 3, 6, 9, 12, 15, 18, and 21). Complexes were recovered on IgG-Sepharose beads and eluted with sample buffer. Eluate (20%) was run on an 8% SDS-PAGE alongside 1/25 of the input SLBP for each binding reaction (lanes 1, 4, 7, 10, 13, 16, and 19). I, input. (C) The different in vitro-translated SLBP truncation or site-specific mutants labeled with [35S]methionine were incubated without import receptor (lanes 2, 6, 10, 14, 18, 22, 26, and 30), with zz-Impα/Impβ (lanes 3, 7, 11, 15, 19, 23, 27, and 31) or with zz-Trn-SR2 (lanes 4, 8, 12, 16, 20, 24, 28, and 32). The samples were subsequently processed as in B.