Abstract
Transcriptional regulatory elements play essential roles in gene expression during animal development and cellular response to environmental signals, but our knowledge of these regions in the human genome is limited despite the availability of the complete genome sequence. Promoters mark the start of every transcript and are an important class of regulatory elements. A large, complex protein structure known as the pre-initiation complex (PIC) is assembled on all active promoters, and the presence of these proteins distinguishes promoters from other sequences in the genome. Using components of the PIC as tags, we isolated promoters directly from human cells as protein-DNA complexes and identified the resulting DNA sequences using genomic tiling microarrays. Our experiments in four human cell lines uncovered 252 PIC-binding sites in 44 semirandomly selected human genomic regions comprising 1% (30 megabase pairs) of the human genome. Nearly 72% of the identified fragments overlap or immediately flank 5′ ends of known cDNA sequences, while the remainder is found in other genomic regions that likely harbor putative promoters of unannotated transcripts. Indeed, molecular analysis of the RNA isolated from one cell line uncovered transcripts initiated from over half of the putative promoter fragments, and transient transfection assays revealed promoter activity for a significant proportion of fragments when they were fused to a luciferase reporter gene. These results demonstrate the specificity of a genome-wide analysis method for mapping transcriptional regulatory elements and also indicate that a small, yet significant number of human genes remains to be discovered.
Promoters are DNA segments located immediately adjacent to the transcriptional start sites of genes (Smale and Kadonaga 2003). They are recognized by both sequence-specific and general transcriptional regulators during transcription initiation, and serve to integrate signals from multiple cellular pathways to deliver specific, highly regulated expression of a gene (Smale and Kadonaga 2003; Hahn 2004). Knowledge of the promoter sequences is essential for understanding the mechanisms of gene regulation during development and physiology. The human genome is composed of a very small fraction of protein-coding sequence (1%-2%) and an overwhelming fraction of intergenic, nonprotein coding sequence (98%). Accurate promoter identification in the human genome has been a great challenge (Zhang 2003).
A complex array of sequence motifs, such as TATA box, INR, DPE, MTE, and BRE, have been described to be associated with promoters; however, these elements are derived from a few select model promoters and are not present in all promoters (Smale and Kadonaga 2003; Hahn 2004). Moreover, these motifs usually have a high degree of degeneracy and are found millions of times in the human genome. As a result, computational identification of gene promoters based on these signature sequences has achieved only limited success.
Annotation of promoters in the human genome has relied primarily on the experimental evidence of 5′ ends of mRNA transcripts, which correspond to the transcription start sites (Ashurst and Collins 2003; Trinklein et al. 2003). A substantial effort has been devoted to cloning and sequencing full-length cDNAs to refine current genome annotation and to define promoters for these genes (Imanishi et al. 2004; The MGC Project Team 2004; Ota et al. 2004). However, these traditional methods are limited by the recovery rate of full-length mRNA sequences, and it is not clear how many more mRNAs remain to be identified. In the DataBase of Transcriptional Start Sites (DBTSS), transcription start sites (TSS) for 8973 human genes have been determined from full-length cDNA sequence information (Suzuki et al. 2004). Similarly in the RefSeq database (Pruitt et al. 2003), which contains a nonredundant set of mRNA coding for a full-length protein, the putative TSS for 17,702 transcripts corresponding to 13,300 genes, inclusive of DBTSS, have been annotated. Given the current estimates of the total human genes at ∼25,000 (including nonprotein-coding genes), promoters for a majority of genes in the human genome remain to be fully defined (International Human Genome Sequencing Consortium 2004).
Here, we use a novel promoter-identification strategy that is independent of our knowledge of mRNA sequences. In this approach, we use genome-wide location analysis (GWLA, Fig. 1A) (Ren et al. 2000; Iyer et al. 2001) to map the genomic binding sites for the transcription machinery associated with transcription start sites. GWLA, also known as ChIP-on-chip, combines chromatin immunoprecipitation (ChIP) of specific protein/DNA complexes from formaldehyde cross-linked cells with genomic DNA microarrays (chip). This technique has been used successfully to map binding sites for nearly all yeast transcription factors in the yeast genome (Lee et al. 2002; Harbison et al. 2004), to identify promoters occupied by human transcription factors in subsets of human promoters (Li et al. 2003; Odom et al. 2004), and more recently, to determine transcription factor binding sites in human chromosomes 21 and 22 (Martone et al. 2003; Cawley et al. 2004; Euskirchen et al. 2004).
Figure 1.
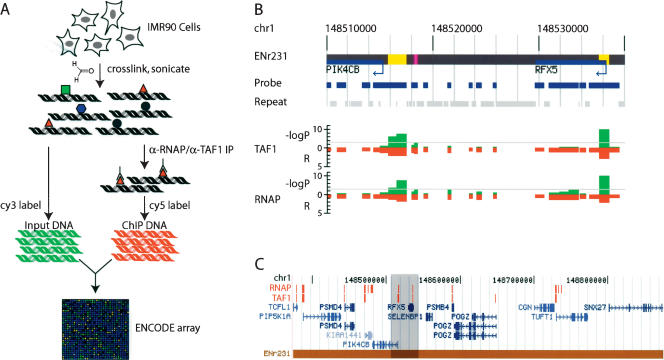
Direct isolation and identification of promoters in the human genome. (A) A schematic of GWLA for mapping RNAP and TAF1-binding sites. Growing cells are cross-linked with formaldehyde, and their nuclei are isolated and sonicated. The resulting chromatin (protein-DNA) complexes are incubated with either anti-RNAP (α-RNAP) or anti-TAF1 (α-TAF1) antibody. The immunoprecipitated DNA is subjected to ligation-mediated PCR, labeled with Cy5 dye, and competitively hybridized to the ENCODE array (described in Supplemental materials) with the Cy3-labeled unenriched chromatin (Li et al. 2003). (B) A typical detailed view of the TAF1- and RNAP-binding data (within the ENCODE region, ENr231). Negative logarithmic P values of enrichment by RNAP or TAF1 ChIP for each probe are plotted in green. Relative enrichment (R) values by RNAP or TAF1 ChIP for each probe fragment are plotted in red on the inverted axis. (C) A representative view of an entire ENCODE locus (ENr231) with annotated RNAP- and TAF1-binding sites that have P values <0.0001, noted in red blocks (The detailed view in B is highlighted in gray).
In eukaryotic cells, active promoters are marked by the assembly of a very large multiprotein structure known as the pre-initiation complex (PIC), which precedes transcription of the corresponding gene (Smale and Kadonaga 2003). Key components of the PIC are the RNA polymerase required for synthesis of RNA and the TFIID complex that recognizes the sequences within the core promoter. Direct biochemical identification of promoters using these promoter-specific protein markers can locate the active promoters in cells independent of the knowledge of mRNA sequence for the transcript. In addition, by sampling different tissues with complementary genome expression profiles, one may reveal the entire set of promoters in a genome.
We describe our initial results of applying the GWLA method to define promoter regions in the human genome to verify feasibility and accuracy of our strategy. We focused on 44 human genomic regions comprising 1% (30 megabase pairs) of the human genome. These regions were chosen by the international ENCODE consortium as an initial effort to map functional elements in the human genome (http://www.genome.gov/10506161, The ENCODE Project Consortium 2004). We designed and manufactured DNA arrays that contained ∼25,000 PCR-amplified DNA fragments with an average length of 600 bp, representing over 92% of the nonrepetitive DNA sequences in the ENCODE regions (see Methods for details). Using GWLA, we isolated the DNA fragments directly associated with the PIC from four different human cell lines, and identified the resulting DNA sequences that fall within the ENCODE regions using the afore-mentioned ENCODE arrays (Fig. 1A). The vast majority of the PIC-binding sites are located at or immediately next to 5′ ends of known genes. However, a significant number of PIC sites is found within intronic or intergenic regions, and may correspond to putative promoters of novel genes. We utilized conventional molecular biology techniques to confirm that a large proportion of these putative promoter regions indeed possess promoter activities and can function as transcription start sites. Our results demonstrate that the genome-wide location-analysis approach can be used to directly map promoter regions in the human genome in the absence of transcript information. We propose that this strategy can also serve to map other types of transcription regulatory elements.
Results
Mapping RNAP-binding sites in the ENCODE regions in IMR90 cells
All class II genes are transcribed by RNA polymerase II (RNAP); thus, association of RNAP would indicate sites of active transcription (Smale and Kadonaga 2003). Moreover, the RNAP found within the initiation site is hypophosphorylated at its C-terminal domain (CTD) and is distinct from the elongating or terminating forms of the enzyme that are hyperphosphorylated (Hahn 2004; Sims III et al. 2004). We therefore used a monoclonal antibody that specifically recognizes the unphosphorylated RNAP CTD to isolate and identify the DNA sequences surrounding the sites of PIC formation (i.e., active promoters) in the human genome (Thompson et al. 1989; Smale and Kadonaga 2003). Using this antibody, we performed genome-wide location analysis experiments with the chromatin isolated from IMR90 human primary fibroblast cells that are derived from a fetal lung tissue. The results from these experiments were analyzed using a previously described error-model (Roberts et al. 2000; Li et al. 2003). From three independent experiments, we combined the enrichment ratios and significance values for each array element to obtain a weighted average of enrichment ratios and P values. The resulting enrichment ratios and log transformed P values for each array element were plotted against their chromosomal positions along with gene annotations. As shown in Figure 1B, a majority of the RNAP-binding sites appear to localize to transcription start sites. Since the average size of the IMR90 chromatin fragments in our experiment is 1.5 Kbp and the average length of the array elements on our ENCODE array is 600 bp, enrichment signals are often detected on neighboring elements as well as on the expected RNAP-binding site, providing additional support for the binding of RNAP to the regions (Fig. 1B). To eliminate the potential redundancy in the identified RNAP-binding sites, we defined an RNAP-binding site as a cluster of neighboring probes that have significant enrichment signals with the P value <0.0001. Using this criterion, we found that RNAP binds to a total of 182 sites (corresponding to 278 individual array elements) (Fig. 1B; Supplemental Fig. S1A; Supplemental Tables S1A, S2A).
Mapping TFIID-binding sites in the ENCODE regions in IMR90 cells
To assess the quality and accuracy of our RNAP GWLA results, we also performed independent GWLA assays to identify genomic sites bound by the general transcription factor TFIID (Supplemental Fig. S1A), which recognizes core promoter motifs and plays a general role in transcriptional initiation in eukaryotic cells (Hahn 2004). For this experiment, we used a monoclonal antibody that specifically recognizes the TAF1 (previously known as TAF250) subunit of the TFIID complex (Ruppert et al. 1993; Espinosa et al. 2003). Using the same analysis method and P value threshold as RNAP, we identified a total of 172 TAF1-binding sites (corresponding to 258 individual array elements on the array) within the ENCODE regions (Supplemental Tables S1, S2).
As expected, there is a strong concordance between RNAP- and TFIID-binding regions in the IMR90 cells. Both complexes are typically colocalized to the annotated promoters, but not to the intergenic regions or transcribed sequences, indicating a high degree of specificity of our promoter mapping method (Fig. 1B,C; Supplemental Fig. S5). Of all of the TFIID-binding regions that we identified, 114 (66%) are also bound by RNAP (P < 0.0001). The remainder appears to be weakly associated with RNAP, as many of these TFIID-bound sites correspond to weak confidence values (0.01 > P > 0.0001), and consequently, are above the threshold cutoff that we have used. Similarly, a majority of the RNAP-binding sites are also associated with TFIID. Some RNAP sites are not associated with TFIID in these cells, as well as a few TFIID sites that are occupied by TFIID, but not RNAP. Many of these TFIID-absent RNAP and RNAP-absent TFIID sites are near the 5′ ends of transcripts and may indicate that these sites represent partially assembled PIC (Supplemental Figs. S3, S4). In addition, some of the TFIID-absent RNAP sites correspond to the transcribed regions downstream of the identified TFIID-binding sites, and may be attributable to the persistence of hypophosphorylated RNAP in these regions.
Promoters can be specifically defined by binding of RNAP and TFIID
To refine and to increase the accuracy of promoter identification, we combined the TFIID- and RNAP-binding information to define active promoters in IMR90 cells by requiring the binding of both TFIID and RNAP (P < 0.001 for RNAP and TAF1, and P < 0.0001 for either RNAP or TAF1). Using this criterion, we identified a total of 118 distinct PIC-binding sites that represent active promoters (corresponding to 164 array elements, Supplemental Table S1C).
Although the accuracy of the 5′-end sequences in the public databases remains uncertain and incomplete, we used the available sequence information to determine whether the identified PIC-binding sites are near the 5′ end of known cDNA sequences. To account for the uncertainty of true 5′-end positions of known transcripts in the public databases, and the relatively low resolution of mapped RNAP- and TAF1-binding sites, we chose an arbitrary distance of 2.5 Kbp as a measure of close proximity between the binding sites and the putative 5′ ends. We first searched for matching sequences in the manually curated RefSeq collection. A significant fraction (58%) of the PIC-binding sites are within 2.5 Kbp of the annotated transcription start sites (Pruitt et al. 2003) (Supplemental Tables S1C, S3A), confirming that our method can identify gene promoters.
The remaining 49 PIC-binding sites (42%) are >2.5 Kbp from any 5′ ends of RefSeq mRNA sequences. To verify that these sequences contain bona fide promoters and transcription-initiation sites, we expanded our search to include any known full-length mRNAs in GenBank (Benson et al. 2003) that may be potential transcripts initiated from these promoters. We compared the location of the 49 putative promoters with the existing mRNA transcript sequences in GenBank and found 27 of the 49 putative promoters to be within 2.5 Kbp of the 5′ end of full-length mRNA transcript reported in GenBank (Supplemental Tables S1C, S2C, S3A). In summary, 81% of the identified PIC-binding sites (87% of individual probes) are within 2.5 Kbp of the 5′ end of RefSeq or GenBank cDNA sequences. These PIC-binding sites contain a promoter for the corresponding transcripts. The remaining 22 PIC-binding sites may contain putative promoters for transcripts that are not yet annotated.
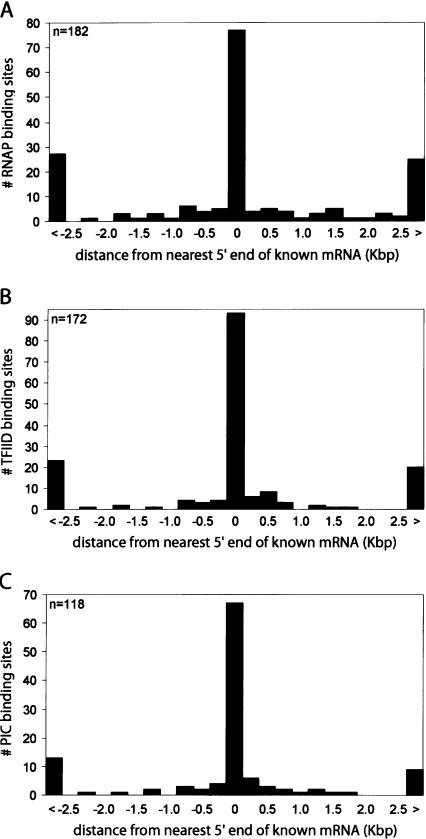
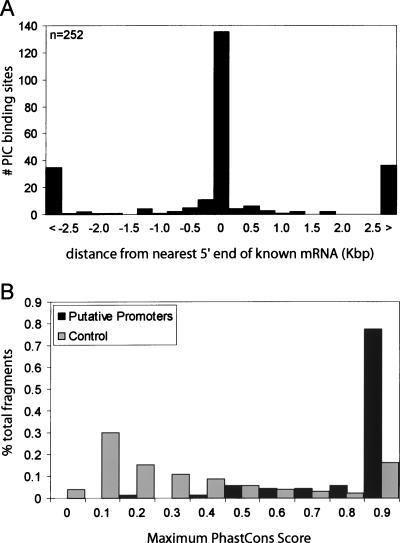
We also analyzed the relative distances of the RNAP sites, TFIID sites, or RNAP/TFIID sites to the 5′ ends of the nearest RefSeq or GenBank transcripts (Supplemental Tables S1A,B,C, S2A,B,C). A vast majority of the RNAP, TAF1, or RNAP/TAF1-binding sites are near the 5′ end of known transcripts (Fig. 2A-C). In contrast, we observed few RNAP- or TAF1-binding sites, and no RNAP/TAF1 sites near the 3′ end of the known transcripts (Supplemental Fig. S4). This result indicates that binding of TAF1 and RNAP serves as a highly reliable marker for gene promoters.
Figure 2.
Summary of identified PIC-binding sites matched to transcripts in IMR90. Histograms plotting relative distance of the RNAP-, TFIID-, or PIC-binding sites to the nearest 5′ ends of full-length GenBank RefSeq transcripts. (A) Distribution (number of RNAP-binding sites, y-axis) of relative distances (in Kbp, x-axis) of the RNAP-binding sites to the nearest 5′ ends of full length mRNA. The first and last bars are counts for those RNAP-binding sites that are >2.5 Kbp upstream (<) or downstream (>) from the nearest 5′ end. (B) Distribution (number of TFIID-binding sites, y-axis) of relative distances (in Kbp, x-axis) of the TFIID-binding sites to the nearest 5′ ends of full-length mRNA. The first and last bars are counts for those TFIID-binding sites that are >2.5 Kbp upstream (<) or downstream (>) from the nearest 5′ end. (C) Distribution (number of PIC-binding sites, y-axis) of relative distances (in Kbp, x-axis) of the PIC-binding sites to the nearest 5′ ends of full-length mRNA. The first and last bars are counts for those PIC-binding sites that are >2.5 Kbp upstream (<) or downstream (>) from the nearest 5′ end.
Novel promoters are active and produce novel transcripts
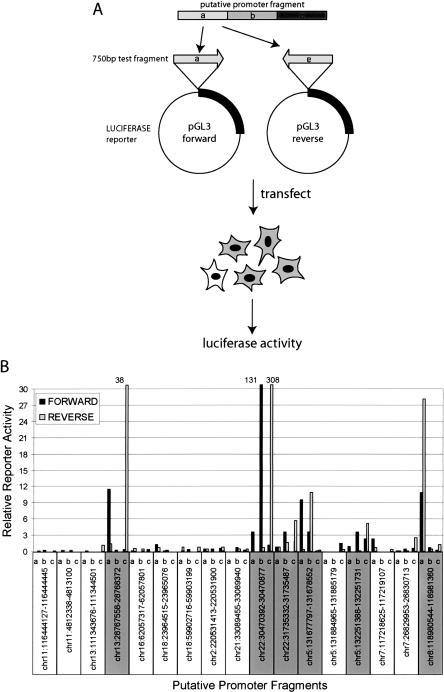
We were not able to assign RNA transcript for 22 PIC-binding sites (Supplemental Tables S1C, S3B). To test whether these sites may correspond to novel promoters for unannotated transcripts, we used a high-throughput reporter assay system and tested whether the 22 putative promoter fragments could drive transcription of a reporter gene in tissue culture cells (Trinklein et al. 2003). We divided each fragment into three DNA fragments of 750 bp and tested the promoter function in both orientations (Fig. 3A). Seventeen PIC-binding sites were successfully cloned and were further tested in transient transfection assays. Six luciferase reporter constructs for each promoter fragment were transfected into a fibrosarcoma cell line, HT1080, which is a similar cell type as IMR90 cells, but could be more easily transfected. The luciferase reporter activity was determined 48 h post-transfection, and was compared with the reporter activities of 89 control DNA fragments randomly selected from the human genome (Trinklein et al. 2003). The activities from at least 14 reporter constructs corresponding to six putative promoter fragments were significantly higher than the controls (three standard deviations over the average reporter activity in control reporter constructs) (Fig. 3B; Supplemental Table S3C,D). Interestingly, many of the putative promoter fragments displayed bidirectional transcriptional activity in the assay system, indicating that they may correspond to divergently transcribed genes. Abundance of such bidirectional promoters in the human genome has been noted previously (Adachi and Lieber 2002; Trinklein et al. 2004).
Figure 3.
Experimental validation of the putative promoters by reporter assays. (A) A schematic of reporter assay used to determine whether the identified putative promoter fragment can support transcription. Each putative promoter fragment was segmented into 750-bp fragments by PCR and cloned into the luciferase reporter construct, pGL3, in either forward or reverse orientations. The resulting reporter constructs were individually transfected into HT1080 cells and the resulting luciferase activity was measured. (B) Reporter activities of 17 putative promoter fragments are shown (remaining five of 22 were not tested). Relative reporter activity was determined by comparing the luciferase activity of the test fragment (Supplemental Table S3C) and the control genomic DNA fragments (Supplemental Table S3D). Promoter fragments with a significant reporter activity (exceeding three times the standard deviation of all control fragments) are highlighted in gray.
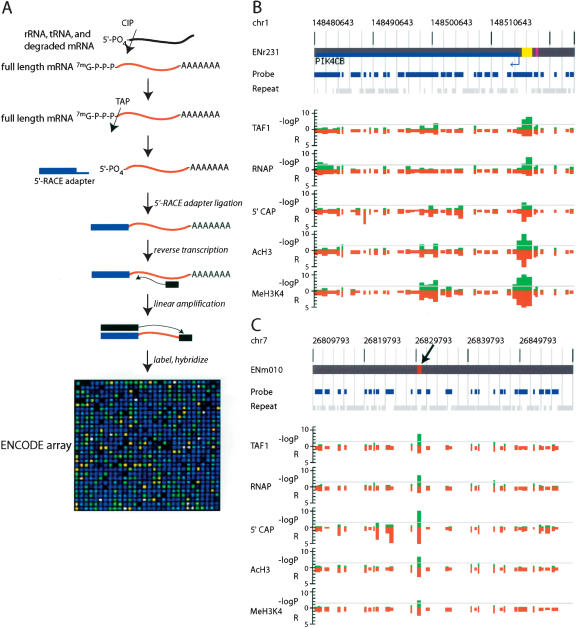
Results from the reporter assays suggest that 35% (6 of 17) of the PIC-binding sites that contain putative promoters indeed possess promoter activity in vivo. The failure to drive reporter expression by the remaining putative promoter fragments may indicate that they are false positives. Alternatively, other technical factors, such as a low sensitivity of the reporter assays or cell type-dependent promoter activities, may have resulted in the negative result. In order to investigate further the identity of these putative promoters, we examined whether any detectable transcript is generated from the putative promoters in IMR90 cells. To this end, we developed an array-based method (RNA ligase-mediated rapid amplification of cDNA ends, ARLM-RACE) to map the 5′ ends of mRNA within the ENCODE regions (Fig. 4A). ARLM-RACE is a modified oligo-capping method (Maruyama and Sugano 1994) (also known as RLM-RACE) that has been adapted to genomic tiling arrays. The method involves selective enrichment of the 5′-capped mRNA sequences by sequential treatment of the RNA sample with calf intestine phosphatase (CIP) and tobacco acid pyrophosphatase (TAP) to remove the 5′ methyl cap and ligation of the 5′RACE adapter oligo to the decapped mRNA. This was followed by linear amplification of the 5′-capped sequences and hybridization of the resulting fragments to the custom-designed ENCODE array to determine the 5′ end of recovered mRNA molecules.
Figure 4.
Experimental validation of the putative promoters by detection of 5′-capped mRNA. (A) A schematic of ARLM-RACE used to map 5′-capped mRNA sequences within the ENCODE regions. (CIP) Calf Intestine Alkaline Phosphatase; (TAP) Tobacco Acid Pyrophosphatase (the decapping enzyme). A detailed procedure is described in the Methods. (B) Detection of the 5′-capped mRNA sequence from and localized acetylated histone H3 (AcH3) and methylated histone H3 lysine residue 4 (MeH3K4) at a known promoter (marked with yellow box). Negative logarithmic P values of enrichment by RNAP ChIP, TAF250 ChIP, ARLM-RACE, Acetyl Histone H3, and Methyl K4 Histone H3 for each probe are plotted in green and relative enrichment (R) values for each probe fragment are plotted in red on the inverted axis. (C) Detection of the 5′-capped mRNA sequence from a previously unknown promoter (marked with magenta box and denoted by an arrow). Negative logarithmic P values of enrichment by RNAP ChIP, TAF250 ChIP, ARLM-RACE, Acetyl Histone H3, and Methyl K4 Histone H3 for each probe are plotted in green and relative enrichment (R) values for each probe fragment are plotted in red on the inverted axis.
A representative ARLM-RACE result is shown in Figure 4, B and C, along with the RNAP and TFIID GWLA results. ARLM-RACE correctly revealed the 5′ end for roughly 40% of the known genes in the ENCODE regions (Fig. 3B). Of the 22 putative promoters, nine were confirmed to have produced 5′-capped transcripts (Fig. 4C; Supplemental Table S3B). Therefore, a large proportion of the putative promoters are indeed true promoters. However, we were not able to detect 5′-capped transcripts for about half of the novel promoters (Supplemental Table S3B; Supplemental Fig. S5). This could be due to a low sensitivity of the current ARLM-RACE protocol. In order to define any transcripts that were produced from these promoters, we performed a traditional oligo-capping method (Maruyama and Sugano 1994) to determine the actual 5′-end sequences of mRNA transcribed from these putative promoters. We designed specific primers for 13 putative promoters based on available gene-prediction models near these putative promoters and cloned the 5′ RLM-RACE products and sequenced their 5′ ends. Six were confirmed to have produced transcripts based on recovery of a specific 5′-end sequence within 2.5 Kbp from the promoter (Supplemental Table S3C).
Together, ARLM-RACE and conventional RACE methods provided confirmation that 12 of the 22 novel promoters are indeed functional promoters in IMR90 cells. Since both the 5′ RACE-based method and the reporter assays verified nearly 50% of the putative promoter fragments, we estimate that approximately half of the 22 putative PIC-binding sites mapped outside of 2.5 Kbp of 5′ ends of RefSeq or GenBank mRNA contain true promoters (Fig. 3B). When combined with the number of PIC-binding sites that matched to known 5′ ends of annotated cDNA sequences, the above results indicate that at least 90% of the PIC-binding sites (107 of the 118 promoters) that we have identified through GWLA experiments correspond to promoters for either known or novel transcripts.
Identified promoters display localized histone H3 acetylation and methylation
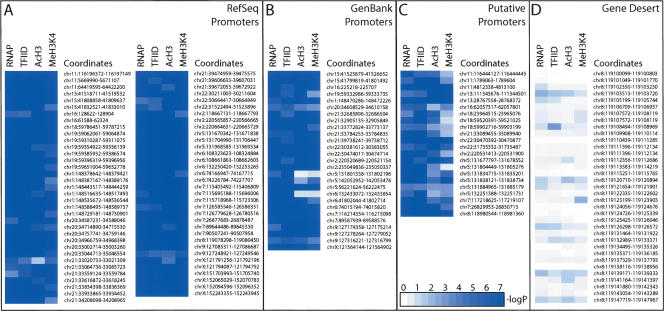
To determine whether the novel promoter fragments exhibit similar characteristics as the known promoters, we examined additional features of these sequences. Specifically, we examined acetylation of histone H3 and methylation of histone H3 at residue 4 lysine, which are known to be associated with gene activation (Jenuwein and Allis 2001; Fischle et al. 2003; Liang et al. 2004; Schneider et al. 2004; Schubeler et al. 2004). Consistent with the previous observations, a highly localized histone H3 acetylation and methylation pattern was observed among known gene promoters (Fig. 5A-C; Supplemental Figure S5), but not in intergenic or heterochromatic regions (Fig. 5D; Supplemental Figs. S1E, S5). Strikingly, 96% of all PIC-binding sites that matched to the 5′ end of known mRNA were associated with either hyperacetylated histone H3 or histone H3 methylated at lysine residue 4 at the confidence level of P < 0.001. All but three of the 22 putative promoter fragments also exhibit histone acetylation and methylation patterns typical of active promoters (Fig. 5C; Supplemental Tables S3B, S3C). Furthermore, all six novel promoter fragments, for which we were not able to obtain their 5′-capped transcript information or confirm their activity by reporter assays, exhibit a localized histone acetylation and methylation pattern. This observation raises the possibility that most of these putative promoter fragments may also drive expression of novel transcripts.
Figure 5.
Summary of histone modification status obtained for each PIC-binding site identified in IMR90 cells. White to blue gradient coloring is used to represent the negative logarithmic P values indicated in each column for RNAP ChIP, TAF250 ChIP, Acetyl Histone H3 (AcH3) ChIP, and Methyl K4 Histone H3 (MeH3K4) ChIP experiment data. (A) RefSeq supported promoters are listed. (B) GenBank supported promoters; (C) putative promoters; (D) transcriptionally inactive, heterochromatic region within ENr321 (gene desert). Actual confidence values for all the promoters are provided in Supplemental Tables S1 and S2. The coloring scale is noted in the bottom inset in C.
Analysis of promoters from different cell types
Since only a small fraction of all promoters actually associate with RNAP and TFIID in each cell type, multiple cell types are needed to identify all promoters in the human genome. To assess the number of cell types needed for comprehensive coverage of human promoters using GWLA analysis, we performed experiments to identify RNAP- and TFIID-binding sites in three additional cell lines, e.g., HCT116, HeLa, and THP1 (Supplemental Figs. S1B,C,D, S6), corresponding to colorectal epithelial, ovarian epithelial, and peripheral blood cell types, respectively. Combining the results from all four cell lines, we were able to identify 252 PIC-binding sites within the ENCODE regions (399 positive probes), representing a 114% increase from IMR90 alone. Analysis of the relative distances of the sites bound by both RNAP and TFIID in all four cell lines to the 5′ ends of nearest RefSeq or GenBank transcripts (Fig. 6A) revealed that a vast majority of the bound sequences are highly localized near the 5′ end of the known transcripts (Fig. 6A). Among these PIC-binding sites, 119 are localized within 2.5 Kbp of the 5′ ends of known transcripts in RefSeq database (Fig. 6A; Supplemental Tables S1, S2, S3D). The remaining 133 fragments are located near the 5′ end of full-length cDNAs in GenBank and, therefore, likely serve as transcription-initiation sites for previously known transcripts (Fig. 6A; Supplemental Table S4). Thus, 72% of 252 PIC-binding sites (or 78% of the 399 positive probes) contain promoters for known transcripts (for examples of alternate promoter usage, see Supplemental Table S1I). The remaining 71 PIC-binding sites most likely contain putative promoters for previously unannotated transcripts in the ENCODE regions (Fig. 6A; Supplemental Table S3E).
Figure 6.
Summary of promoters identified in IMR90, HCT116, HeLa, and THP1 cells. (A) Distribution (number of PIC-binding sites in all four cell lines, y-axis) of relative distance (in Kbp, x-axis) of the PIC-binding sites to the nearest 5′ ends of full-length GenBank mRNA. The first and last bars are counts for those PIC-binding sites that are >2.5 Kbp upstream (<) or downstream (>) from the nearest 5′ end. (B) Conservation analysis of 71 putative promoters identified in four cell lines (in dark gray bars) and control genomic fragments (in light gray bars). The x-axis represents conservation score, PhastCons, and the y-axis represents the percentage of all putative promoters (or the control genomic fragments) with the corresponding PhastCons score.
We have examined the conservation of the regions surrounding the putative promoters identified in all four cell types. By analyzing the putative promoter sequences and the 2.5-Kbp surrounding sequences, we found that 55 (77%) of the 71 putative promoters contain highly conserved regions with a maximum PhastCons score of 0.9 or greater. By comparison, only 16% of randomly selected fragments within the ENCODE region had a maximum PhastCons score of 0.9 (Siepel and Haussler 2003) (Fig. 6B; Supplemental Table S1H). Our results indicate that a majority of putative promoter sequences are evolutionarily conserved, consistent with their function as promoters.
Discussion
We have outlined a strategy that can accurately define active promoters in the genome. This promoter-mapping strategy is unbiased and independent of our knowledge of mRNA sequences. To demonstrate the feasibility, efficiency, and specificity of this strategy, we have investigated four human cell lines and identified a total of 252 genomic sites in the 1% human ENCODE regions that are bound by PIC and most likely function as promoters in at least one of the four cell types examined. A vast majority of these sites corresponds to the 5′ end of known transcripts, while nearly 28% of these sites may represent putative promoters for not yet annotated transcripts. By using reporter assays and 5′ RACE-based methods, we further verified that nearly half of the putative promoters identified in one cell line contain bona fide promoters that can initiate transcription in vivo. Therefore, using the information we have obtained from this method, we can define promoters and uncover evidence of novel genes.
Efficiency and accuracy of our promoter-mapping strategy
The most conservative estimate of specificity of our method is between 88% and 91%, based on results of two independent verification approaches applied to the 22 putative promoters identified in IMR90 cells. However, the actual specificity of our method is likely higher, since both reporter assays and RACE-based methods may be limited by sensitivity. In addition, virtually all of the putative promoters displayed a highly localized histone-modification pattern that is typical of all active promoters.
The fact that we require the binding of both RNAP and TFIID as criteria for promoters may have led to the elimination of some promoters that show partial binding of either TAF1 or RNAP from our final list, and resulted in a relatively moderate sensitivity. The number of RefSeq genes in the ENCODE regions whose transcript was detected by Affymetrix transcriptional profiling experiments is 149. Of these, the 5′ ends of 75 genes overlap or are near the PIC-binding sites that we have mapped. For the remaining 74 genes, we did not detect PIC-binding sites near their 5′ ends (Supplemental Table 3H). Further experiments are required to determine the biological basis for the absence of PIC binding to the promoters of those genes whose corresponding transcripts are present. It is possible that these promoters are transiently or weakly associated with the PIC, thus making the detection by GWLA difficult. Conversely, it is also possible that these transcripts are highly stabilized, and actual transcription of these genes rarely occurs. In any case, the most conservative estimate of sensitivity of our method is slightly over 50%. Analysis of multiple cell types is going to increase the coverage of promoter identified using this approach. Our analysis of four distinct cell lines led to the identification of twice as many PIC-binding sites as those that would be identified in a single cell line. A full coverage of all human promoters may be achieved by expanding the panel of different tissues and cell lines.
Another limitation of our current approach is the final resolution of the binding sites. Due to the large size of fragmented chromatin (around 1.5-Kbp fragments) and the size of each probe on our ENCODE microarrays, the current resolution is limited to about a 1-2 Kbp fragment surrounding the 50-100-bp expected footprint of PIC. By increasing the sonication time and including enzymatic treatment, the average chromatin fragment size can be reduced to 500 bp or less, which may lead to a slight increase in resolution. A more general approach to enhance the resolution of PIC-binding sites may be to use genome tiling arrays composed of short oligonucleotide probes that cover the genome at a much higher density. With the development of better analysis algorithms and increased density of the genome tiling arrays, we expect to refine the resolution of the PIC binding sites and may even be able to locate the actual footprint of these factors.
Promoter mapping may lead to identification of novel genes
Current gene annotation of the human genome is heavily biased toward protein-coding genes. However, recent studies have suggested the existence of a large number of noncoding RNA genes in the genome (Kapranov et al. 2002; Kampa et al. 2004; Ota et al. 2004). These noncoding RNA genes could correspond to miRNA or other noncoding RNA, such as Xist, that regulate gene expression (Bartel 2004). Unlike the protein-coding mRNAs, these noncoding RNAs may be extensively processed, and identification of their corresponding genes may be difficult by conventional molecular biology techniques. Our approach to map gene promoters does not rely on our knowledge of mRNA isolation and detection; therefore, it can be used to discover these genes. Using this strategy, we can profile the entire promoter usage for a given cell systematically and define the transcripts that are generated from these promoters. Furthermore, this strategy represents a directed method for finding and cloning those genes that have escaped traditional methods.
Genome-wide mapping of functional elements in the human genome
The approach described here represents a general approach applicable to mapping other transcriptional regulatory elements in the genome such as enhancers, silencers, and insulators. Like promoters, these functional elements are also associated with transcription factors and specific histone-modification patterns (Brivanlou and Darnell Jr. 2002). Genome-wide location analysis of DNA associated with various histone modifications, transcription factors, and/or their cofactors will allow us to systematically identify regulatory elements in the human genome and decipher how the entire genome is expressed to give rise to the form and function of individual cells and tissues.
Methods
Encode array design
Repeat-masked sequences for the 44 ENCODE regions were downloaded from UCSC Genome Browser using human genome assembly hg16. The repeat sequences constituted 13,477,514 bp, or 45.06% of the ENCODE regions. The remaining nonrepetitive sequences were further processed for the array. Small fragments shorter than 200 bp were excluded from analysis and large fragments longer than 1000 bp were split, so that each resulting fragment is <1000 bp. The total number of bases represented in the array is 15,112,006 bp, or 50.53% of the ENCODE regions, or 92% of nonrepetitive DNA. A total of 25,162 nonrepetitive fragments were selected, with an average fragment size of 600 bp, for PCR amplification. A total of 24,537 pairs of primers were successfully designed using the Primer3 program (Rozen and Skaletsky 2000) and synthesized commercially (Qiagen). These specific primers were used to PCR amplify the DNA fragments within the ENCODE regions. We obtained a >96% PCR success rate in the initial round of PCR. For the failed PCR amplifications, the primers were resynthesized, and a second round of PCR was performed, resulting in the final PCR success rate of >98%. After PCR amplification, we purified each DNA fragment by Perfectprep PCR cleanup kit (Eppendorf AG), verified the product by agarose gel electrophoresis (E-Gel, Invitrogen), and then spotted the purified DNA to GAPSII glass slides (Corning) using a contact printer (Genomic Solutions). The spotted slides were UV cross-linked, and stored under vacuum until use.
GWLA
Chromatin immunoprecipitation was performed as described previously (Li et al. 2003). Briefly, 2 mg of sonicated chromatin (OD260) was incubated with 10 μg of mouse monoclonal RNAP (clone 8WG16, Abcam) or TAF1 antibody (catalog #sc-735, Santa Cruz Biotechnology) coupled to the sheep anti-mouse IgG magnetic beads (Dynal Biotech). The magnetic beads were washed eight times with RIPA buffer containing 50 mM Hepes (pH 8.0), 1 mM EDTA, 1% NP-40, 0.7% DOC, and 0.5 M LiCl, supplemented with Complete protease inhibitors (Roche Applied Science), and washed once with TE (10 mM Tris at pH 8.0, 1 mM EDTA). After washing, the bound DNA was eluted by heating the beads to 65°C in elution buffer (10 mM Tris at pH 8.0, 1 mM EDTA, and 1% SDS). The eluted DNA was incubated at 65°C for 12 h more to reverse the cross-links. Following incubation, the immunoprecipitated DNA was treated sequentially with Proteinase K and RNase A, and was desalted using the QIAquick PCR purification kit (Qiagen). The purified DNA was blunt ended using T4 polymerase (New England Biolabs) and ligated to the linkers (oJW102, 5′-GCGGTGACCCGGGAGATCTGAATTC-3′, and oJW103, 5′-GAATTCAGATC-3′). The ligated DNA was subjected to ligation-mediated PCR, labeled with Cy3 and Cy5 dCTP using a BioPrime DNA labeling kit (Invitrogen), and hybridized to the ENCODE microarray. The hybridized slides were washed and scanned using a Genepix 4000B scanner (Axon Instruments) as described previously (Li et al. 2003).
Reporter assays
PCR amplification, cloning, transfection, and luciferase assays were performed as described previously (Trinklein et al. 2003).
ARLM-RACE
An RNA ligase-mediated rapid amplification of cDNA ends (or oligo-capping method) (Maruyama and Sugano 1994) was performed according to the protocol provided by FirstChoice RLM-RACE kit (Ambion) with the following modifications: 10 μg of total RNA (extracted from IMR90 cells using Trizol reagent, Invitrogen) were treated with 10 U of Calf Intestine Alkaline Phosphatase (CIP) in 20 μL for 1 h at 37°C. The phosphatase-treated RNA was extracted with acid phenol:chloroform and precipitated with ammonium acetate and ethanol. The RNA pellet was resuspended in dH2O and treated with 10 U of Tobacco Acid Pyrophosphatase (TAP) for 1 h at 37°C. The decapped RNA was ligated to the 5′RACE adapter (provided in the kit) using T4 RNA ligase. The ligated RNA was reverse transcribed using M-MLV Reverse Transcriptase and random decamers. The resulting cDNA was subjected to linear amplification by PCR using 5′RACE adapter primer with 35 thermal cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec. The amplified, 5′ cap-enriched DNA was treated with 10 μg of RNase A and 10 μg RNase H for 1 h at 37°C. The cDNA was purified using the QIAquick PCR purification kit (Qiagen) and labeled with Cy5 dCTP using the BioPrimer DNA-labeling kit (Invitrogen) and hybridized with 2 μg of labeled DNA to the ENCODE microarray along with 2 μg of the Cy3-labeled genomic DNA from IMR90 cells for 16 h at 60°C. The hybridized slides were washed and scanned using a Genepix 4000B scanner as described previously (Li et al. 2003).
Data analysis
The raw microarray data are available in GEO (Submissions GPL1454, GSE1778, GSM30733-GSM30789). Analysis of microarray-scanning images was performed according to published protocols with modifications (Li et al. 2003). Data from independent replicate experiments were combined. An average enrichment ratio was calculated for each DNA species on the array from three replicate data sets. The binding of a factor to DNA was deemed significant if the average P value was <0.0001 and the P value of the individual replicate was <0.01 in at least two replicate experiments. By using these criteria, no DNA achieved significance in control experiments where input DNA was compared with the same DNA.
Identified binding sites corresponding to ENCODE array sequences were merged into clusters if within 1.2 Kbp of each other. Probes on the ENCODE array that showed >70% sequence identity to other sequences in the ENCODE regions longer than 100 bp were excluded from the analysis. Human RefSeq transcripts downloaded from UCSC Genome Browser (Karolchik et al. 2003). were used to classify clusters as representing known or novel predicted promoters. A total of 416 RefSeq transcripts with “Provisional”, “Reviewed”, or “Validated” status overlap the ENCODE regions. Clusters within 2.5 Kbp of the annotated 5′ ends of these RefSeq transcripts were classified as known promoters. Clusters outside 2.5 Kbp of the RefSeq transcripts were analyzed further by examining whether they are within 2.5 Kbp of the UCSC-annotated 5′ ends of human mRNA transcripts from GenBank (Karolchik et al. 2003). Remaining clusters outside 2.5 Kbp of annotated 5′ ends of RefSeq and GenBank mRNA transcripts were determined as novel. Analysis of individual probes that are bound by RNAP and/or TAF1 are included in the Supplemental information.
Acknowledgments
We thank Bruce Hamilton and Webster Cavenee for critical reading of the manuscript and the UCSD BioGem Facility for access to their equipment. This research was supported by the Ruth L. Kirschstein National Research Service Award 1F32CA108313 (T.H.K.), Ford Foundation Predoctoral Fellowship (L.O.B.), and grant 1U01HG003151 from the National Human Genome Research Institute (B.R.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3430605. Article published online before print in May 2005.
References
- Adachi, N. and Lieber, M.R. 2002. Bidirectional gene organization: A common architectural feature of the human genome. Cell 109: 807-809. [DOI] [PubMed] [Google Scholar]
- Ashurst, J.L. and Collins, J.E. 2003. Gene annotation: Prediction and testing. Annu. Rev. Genomics Hum. Genet. 4: 69-88. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [DOI] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., and Wheeler, D.L. 2003. GenBank. Nucleic Acids Res. 31: 23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivanlou, A.H. and Darnell Jr., J.E. 2002. Signal transduction and the control of gene expression. Science 295: 813-818. [DOI] [PubMed] [Google Scholar]
- Cawley, S., Bekiranov, S., Ng, H.H., Kapranov, P., Sekinger, E.A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A.J., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499-509. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306: 636-640. [DOI] [PubMed] [Google Scholar]
- Espinosa, J.M., Verdun, R.E., and Emerson, B.M. 2003. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol. Cell 12: 1015-1027. [DOI] [PubMed] [Google Scholar]
- Euskirchen, G., Royce, T.E., Bertone, P., Martone, R., Rinn, J.L., Nelson, F.K., Sayward, F., Luscombe, N.M., Miller, P., Gerstein, M., et al. 2004. CREB binds to multiple loci on human chromosome 22. Mol. Cell. Biol. 24: 3804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle, W., Wang, Y., and Allis, C.D. 2003. Binary switches and modification cassettes in histone biology and beyond. Nature 425: 475-479. [DOI] [PubMed] [Google Scholar]
- Hahn, S. 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11: 394-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison, C.T., Gordon, D.B., Lee, T.I., Rinaldi, N.J., Macisaac, K.D., Danford, T.W., Hannett, N.M., Tagne, J.B., Reynolds, D.B., Yoo, J., et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi, T., Itoh, T., Suzuki, Y., O'Donovan, C., Fukuchi, S., Koyanagi, K.O., Barrero, R.A., Tamura, T., Yamaguchi-Kabata, Y., Tanino, M., et al. 2004. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2: 856-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931-945. [DOI] [PubMed] [Google Scholar]
- Iyer, V.R., Horak, C.E., Scafe, C.S., Botstein, D., Snyder, M., and Brown, P.O. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409: 533-538. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T. and Allis, C.D. 2001. Translating the histone code. Science 293: 1074-1080. [DOI] [PubMed] [Google Scholar]
- Kampa, D., Cheng, J., Kapranov, P., Yamanaka, M., Brubaker, S., Cawley, S., Drenkow, J., Piccolboni, A., Bekiranov, S., Helt, G., et al. 2004. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 14: 331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916-919. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31: 51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.I., Rinaldi, N.J., Robert, F., Odom, D.T., Bar-Joseph, Z., Gerber, G.K., Hannett, N.M., Harbison, C.T., Thompson, C.M., Simon, I., et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799-804. [DOI] [PubMed] [Google Scholar]
- Li, Z., Van Calcar, S., Qu, C., Cavenee, W.K., Zhang, M.Q., and Ren, B. 2003. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. 100: 8164-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, G., Lin, J.C., Wei, V., Yoo, C., Cheng, J.C., Nguyen, C.T., Weisenberger, D.J., Egger, G., Takai, D., Gonzales, F.A., et al. 2004. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. 101: 7357-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone, R., Euskirchen, G., Bertone, P., Hartman, S., Royce, T.E., Luscombe, N.M., Rinn, J.L., Nelson, F.K., Miller, P., Gerstein, M., et al. 2003. Distribution of NF-κB-binding sites across human chromosome 22. Proc. Natl. Acad. Sci. 100: 12247-12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, K. and Sugano, S. 1994. Oligo-capping: A simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138: 171-174. [DOI] [PubMed] [Google Scholar]
- The MGC Project Team. 2004. The Status, quality and expansion of the NIH full-length cDNA project: The Mammalian Genome Collection (MGC). Genome Res. 14: 2121-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom, D.T., Zizlsperger, N., Gordon, D.B., Bell, G.W., Rinaldi, N.J., Murray, H.L., Volkert, T.L., Schreiber, J., Rolfe, P.A., Gifford, D.K., et al. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303: 1378-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota, T., Suzuki, Y., Nishikawa, T., Otsuki, T., Sugiyama, T., Irie, R., Wakamatsu, A., Hayashi, K., Sato, H., Nagai, K., et al. 2004. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36: 40-45. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D., Tatusova, T., and Maglott, D.R. 2003. NCBI Reference Sequence project: Update and current status. Nucleic Acids Res. 31: 34-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, B., Robert, F., Wyrick, J.J., Aparicio, O., Jennings, E.G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306-2309. [DOI] [PubMed] [Google Scholar]
- Roberts, C.J., Nelson, B., Marton, M.J., Stoughton, R., Meyer, M.R., Bennett, H.A., He, Y.D., Dai, H., Walker, W.L., Hughes, T.R., et al. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287: 873-880. [DOI] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365-386. [DOI] [PubMed] [Google Scholar]
- Ruppert, S., Wang, E.H., and Tjian, R. 1993. Cloning and expression of human TAFII250: A TBP-associated factor implicated in cell-cycle regulation. Nature 362: 175-179. [DOI] [PubMed] [Google Scholar]
- Schneider, R., Bannister, A.J., Myers, F.A., Thorne, A.W., Crane-Robinson, C., and Kouzarides, T. 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell. Biol. 6: 73-77. [DOI] [PubMed] [Google Scholar]
- Schubeler, D., MacAlpine, D.M., Scalzo, D., Wirbelauer, C., Kooperberg, C., Van Leeuwen, F., Gottschling, D.E., O'Neill, L.P., Turner, B.M., Delrow, J., et al. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes & Dev. 18: 1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel, A. and Haussler, D. 2003. Combining phylogenetic and hidden Markov models in biosequence analysis. In Proceedings of the Seventh Annual International Conference on Computational Molecular Biology (RECOMB 2003), pp. 277-286. [DOI] [PubMed]
- Sims III, R.J., Mandal, S.S., and Reinberg, D. 2004. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell. Biol. 16: 263-271. [DOI] [PubMed] [Google Scholar]
- Smale, S.T. and Kadonaga, J.T. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72: 449-479. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Yamashita, R., Sugano, S., and Nakai, K. 2004. DBTSS, DataBase of transcriptional start sites: Progress report 2004. Nucleic Acids Res. 32: D78-D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, N.E., Steinberg, T.H., Aronson, D.B., and Burgess, R.R. 1989. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J. Biol. Chem. 264: 11511-11520. [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.J., Saldanha, A.J., and Myers, R.M. 2003. Identification and functional analysis of human transcriptional promoters. Genome Res. 13: 308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.F., Hartman, S.J., Schroeder, D.I., Otillar, R.P., and Myers, R.M. 2004. An abundance of bidirectional promoters in the human genome. Genome Res. 14: 62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M.Q. 2003. Prediction, annotation, and analysis of human promoters. Cold Spring Harbor Symp. Quant. Biol. 68: 217-225. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu/; the UCSC Genome Browser.
- http://www.genome.gov/10506161; The ENCODE Project Consortium 2004.