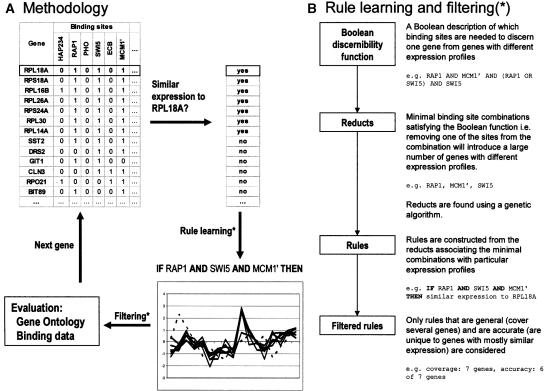
Figure 1.
A schematic description of the method and the rule learning algorithm. (A) Rules are induced from one gene at a time by first identifying similarly expressed genes and then by learning minimal binding-site combinations unique to these coexpressed genes. Filtered rules are finally evaluated using Gene Ontology and binding data by Lee et al. (2002). (B) The rule learning algorithm starts by building a Boolean function describing which binding sites are needed to discern one gene from genes with different expression profiles. This discernibility function is then simplified using a genetic algorithm in order to find minimal binding-site combinations (reducts) satisfying the function. Rules are constructed from the minimal combinations and filtered using accuracy and coverage. The examples given in B are constructed from the small table in A. The obtained reduct (RAP1, MCM1′, SWI5) is the minimal combination needed to discern RPL18A from genes with a different expression. Note that the set of similarly expressed genes in A is indiscernible from the differentially expressed gene SST2 with respect to the binding-site data. The set is thus said to be rough, and the resulting rule has an accuracy that is <1.