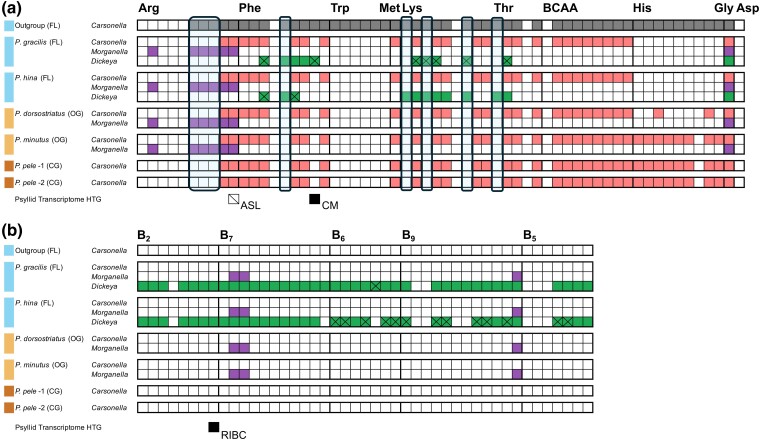
Fig. 5.
Metabolic reconstructions of putative long-term symbionts of Pariaconus psyllid species. Matrices of gene presence, absence, or inactivation in assembled Pariaconus symbiont genomes and an outgroup from same psyllid family (B. cockerelli) for several critical a) amino acid and b) B vitamin biosynthesis pathways. Gene absences (not encoded) are white boxes and gene presence (encoded) are colored boxes corresponding to the Carsonella-, Morganella- (Ca. Makana arginalis), and Dickeya-like (Ca. Malihini olakiniferum) symbiont assemblies. Boxes with an “X” indicate inactivated pseudogene copies. The presence of horizontally transferred genes/transcripts from P. gracilis and P. montgomeri for homologs and/or same enzymes in corresponding symbiont pathways are indicated as detected (black box) or not detected (outlined box with cross).