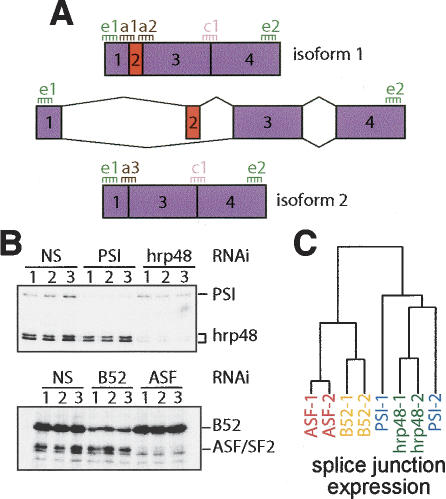
Figure 1.
Experimental design and clustering results. (A) Thirty-six-mer probes were selected for all alternatively spliced junctions from the GadFly 3.2 Drosophila genome annotation (“a” probes). For each gene, two exonic probes were selected from regions common to all isoforms to gauge total gene expression (“e” probes). Up to two constant constitutive junction probes were also selected (“c” probes). (B) Immunoblot analysis confirmed effective RNAi knockdown of the hnRNP proteins PSI and hrp48 and of the SR proteins B52/SRp55 and dASF/SF2. (C) Hierarchical clustering using average log expression ratios from all splice junction probes was performed to assess the global affects of biological replicates of each splicing factor RNAi knockdown and to compare between splicing factors. This analysis indicates that the dASF/SF2, B52/SRp55, and hrp48 experiments produce a characteristic splicing response. The PSI results, however, were more variable (see text). The global splicing response to dASF/SF2 or B52/SRp55 knockdown includes more similarities than either does to hrp48 or PSI knockdown.