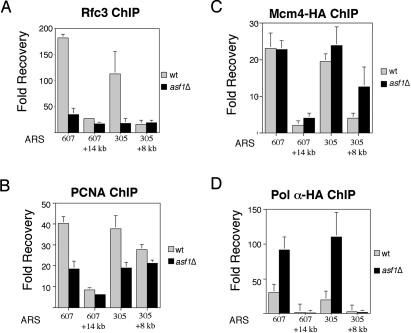
Figure 4.
Differential replication fork protein stability at stalled forks in asf1Δ mutant cells. (A) Rfc3 localization to stalled replication forks requires Asf1. ChIP experiments were performed as described in Figure 3 except that the amount of Rfc3 associated with early origins was assayed using a polyclonal antibody. The amount of DNA recovered from wild-type (WT) and asf1Δ extracts is graphed relative to the amount recovered in the normal rabbit sera control for the early firing origins ARS607 and ARS305, as well as two unreplicated regions 14 kb away from ARS607 (ARS607 + 14 kb) and 8 kb away from ARS305 (ARS305 + 8). (B) PCNA localization to stalled replication forks is reduced in asf1Δ mutant cells. ChIP experiments were preformed and analyzed as described in A except that a polyclonal antibody raised against yeast PCNA was used. (C) Mcm4 localizes to stalled replication forks in the absence of Asf1. ChIP experiments were performed as described above and the amount of origin DNA that coprecipitated with HA-tagged Mcm4 (Mcm4-HA) is graphed relative to the untagged control. (D) Pol α association with stalled replication forks increases in the absence of Asf1. ChIP experiments were performed as described above and the amount of origin DNA that coprecipitated with HA-tagged Pol α (Pol α-HA) is graphed relative to the untagged control.