Figure 3.
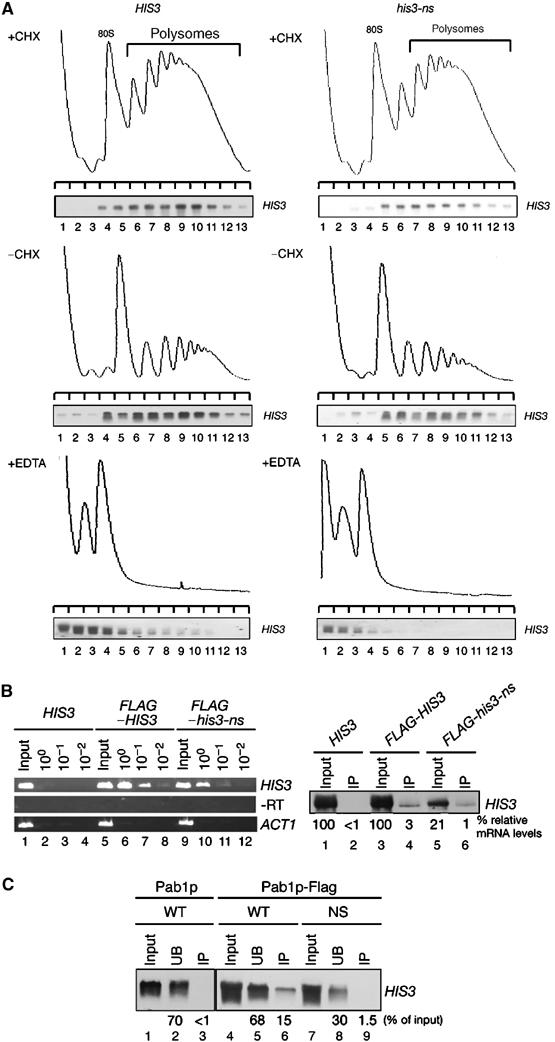
Nonstop mRNAs are associated with ribosomes. (A) W303 cells harboring pIT709 (pHIS3-His6, left panels) or pIT711 (phis3-His6-ns, right panels) were grown on SC-Leu medium. Cell extracts were prepared in the presence (top panels) or absence of (middle panels) CHX. Cell extracts were resolved by velocity sedimentation on 10–50% sucrose gradients. RNA samples prepared from the indicated fractions were analyzed by Northern blotting. Polysome analysis performed in the presence of 30 mM EDTA to separate ribosome subunits is shown in bottom panels. (B) Left: W303 cells harboring the indicated plasmids were grown on SG-Ura medium and cell extracts were prepared in the presence of CHX for affinity purification. RNA samples prepared from purified samples were subjected to RT reaction and cDNA was amplified by PCR with the series of dilutions indicated. Lane 1–4: pIT765 (pGAL1p-HIS3-His6, HIS3); lanes 5–8: pIT826 (pGAL1p-FLAG-HIS3, FLAG-HIS3); lanes 9–12: pIT827 (pGAL1p-FLAG-his3-ns, FLAG-his3-ns). Right: HIS3 mRNAs in samples prepared from total extract or purified samples were detected with Northern blot analysis. (C) W303 cells harboring pIT765 (pGAL1p-HIS3-His6, WT), YIT874 (pab1-FLAGHA TRP1) cells containing pIT765 (pGAL1p-HIS3-His6, WT) or pIT766 (pGAL1p-his3-His6-ns, NS) were grown on SG-Ura medium. An amount of cell extracts equivalent to 40 A260 was used for affinity purification. RNA samples were prepared from cell extracts (lanes 1, 4 and 7), unbound fractions (lanes 2, 5 and 8) and purified fractions (lanes 3, 6 and 9), and analyzed by Northern blotting.
