Figure 7.
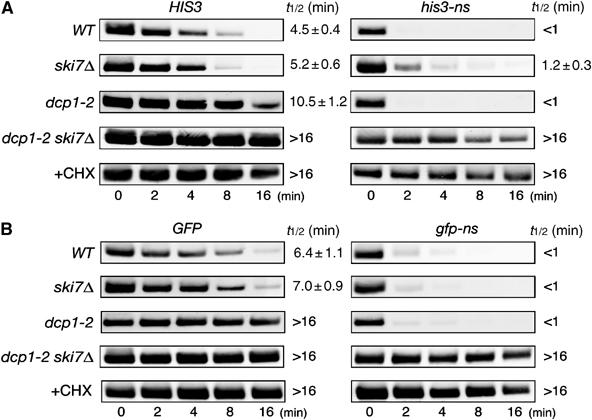
The 5′ → 3′ decay pathway is accelerated for NSD in the absence of Ski7p. (A) Yeast strains (WT, W303; ski7Δ, YS002; dcp1-2, YS088; dcp1-2 ski7Δ, YS091) were transformed with pIT765 (pGAL1p-HIS3-His6, HIS3) or pIT766 (pGAL1p-his3-His6-ns, nonstopHIS3). Cells were grown in SG-Ura. At the beginning of the experiment, glucose was added to inhibit transcription from the GAL1 promoter and samples were harvested at the indicated times. For analysis of mRNA decay in the YS088 (dcp1-2) or YS091 (dcp1-2 ski7Δ) mutant, transcription from the GAL1 promoter was inhibited after incubation at 37°C for 90 min. RNA samples subjected to agarose gel electrophoresis were analyzed by Northern blotting with DIG-labeled HIS3 probe. Where indicated, CHX was added to inhibit translation elongation. The half-lives (t1/2; min) are shown as the mean values±standard deviations (s.d.), which are obtained from at least three independent experiments. (B) Yeast strains (WT, W303; ski7Δ, YS002; dcp1-2, YS088; dcp1-2 ski7Δ, YS091) were transformed with pIT859 (pGAL1p-GFP, GFP) or pIT860 (pGAL1p-gfp-ns, nonstopGFP). Transcriptional repression and hybridization were performed as described for (A), but DIG-labeled GFP probe was used.
