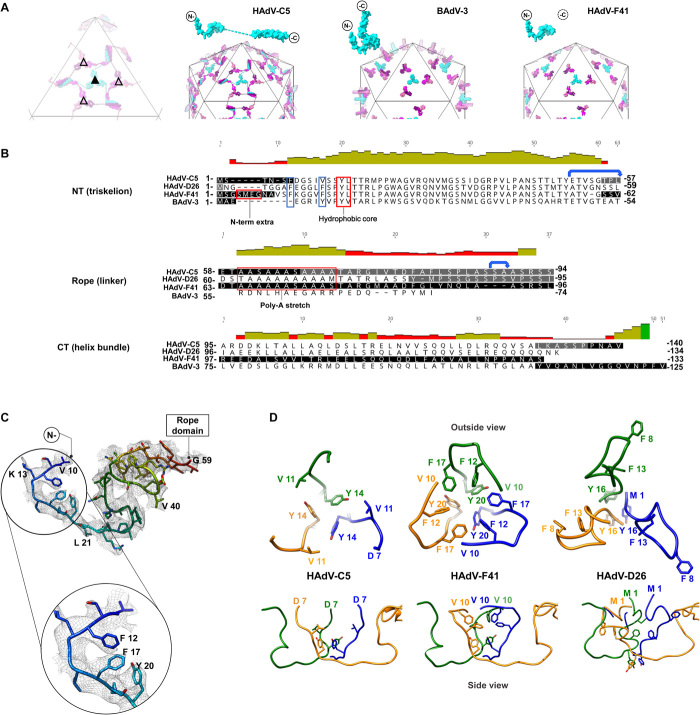
Fig. 3. Protein IX triskelion structure.
(A) Schematics showing the organization of protein IX in HAdV-C5, BAdV-3, and HAdV-F41, where only the N-terminal domain is ordered. Triskelions located at the I3 symmetry axis (filled triangle) are in cyan, and those located at the L3 axis (empty triangle) are in several shades of pink. Dashed lines: rope domains in HAdV-C5 One IX monomer is depicted in cyan on top of each schematic. (B) Structure-based sequence alignment of protein IX in HAdV-C5, HAdV-D26, HAdV-F41, and BAdV-3. Black text: residues modeled in the different structures. White text on black background: regions not modeled. Gray background: regions modeled, but not in all independent copies of IX. Red boxes: N-terminal extra residues in HAdV-F41, triskelion hydrophobic core, and poly-Ala stretch (absent in BAdV-3). Blue boxes highlight the two Phe residues discussed in the text. Blue arrows: the two flexible bends. (C) N-terminal domain of HAdV-F41 protein IX rainbow colored from blue (N terminus) to red (last traced residue), with the density map in gray mesh and a zoom into the hydrophobic residues discussed in the text. (D) Comparison of the triskelions in HAdV-C5 (PDB ID: 6B1T), HAdV-F41, and HAdV-D26 (PDB ID: 5TX1). Top row: triskelions as seen from outside the capsid. Bottom: a view rotated by 90°, so that the hexon shell surface would be located at the bottom.