Figure 7.
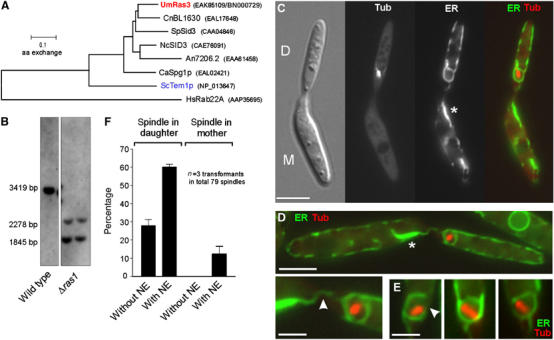
The role of UmRas3, a Temp1-like GTPase in ‘open mitosis'. (A) The genome of U. maydis contains a Ras3-like GTPase that has remarkable similarity to a predicted gene in the pathogenic basidiomycete C. neoformans. The tree is based on a comparison of ∼190 amino acids of the core region of all predicted proteins. Um: Ustilago maydis; Cn: Cryptococcus neoformans; Sp: Schizosaccharomyces pombe; Nc: Neurospora crassa; An: Aspergillus nidulans; Ca: Candida albicans; Sc: Saccharomyces cerevisiae; Hs: Homo sapiens. Accession numbers are given in parentheses. (B) Southern blot confirming that ras3 is deleted. After digestion with BsiWI, wild-type genomic DNA contains a fragment of 3419 bp that is split into smaller fragments in ras3 null mutants. (C) Δras3 mutants show defects neither in morphology nor in cytokinesis. Spindles are positioned correctly within the bud (Tub: CFP-α-tubulin), but are often surrounded by ER (ER-YFP), suggesting that they fail to remove the envelope. Membrane accumulations are found in the mother cell (asterisk), indicating that parts of the envelope are left behind. Bar: 5 μm. (D) The NE often has contact with the membrane accumulation in the mother cell (arrowhead in detail). Bars: 5 and 2 μm. (E) Correctly positioned spindles are often surrounded by a partially opened envelope (arrowhead; see also Table I). (F) Quantitative analysis reveals that Δras3 mutants have only minor defects in spindle positioning (‘spindle in mother'), but ∼60% of the spindle in the daughter bud are surrounded by ER (‘Spindle in daughter, with NE'). Note that all spindles in control strains are in the first category (‘Spindle in daughter, without NE'; see Figure 6A).
