Figure 1.
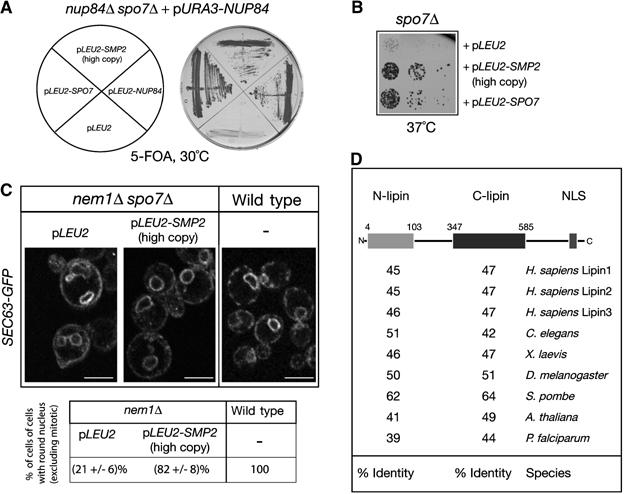
Functional interaction between NEM1–SPO7 and SMP2. (A) Identification of SMP2 as a high-copy number suppressor of the nup84Δ spo7Δ synthetic lethal mutant. The nup84Δ spo7Δ double deletion strain carrying a centromeric vector expressing NUP84 was transformed with the indicated plasmids. Transformants were grown on plates containing 5-FOA for 3 days. (B) SMP2 rescues the temperature-sensitive growth defect of spo7Δ cells. Spo7Δ cells transformed with the indicated plasmids were diluted in YEPD, spotted onto selective (-Leu) plates and grown at 37°C for 2 days. (C) Overexpression of SMP2 suppresses the nuclear membrane proliferation of nem1Δ spo7Δ cells. Upper panel: the nem1Δ spo7Δ mutant or the isogenic wild-type strain, expressing the ER marker Sec63-GFP were transformed with the indicated plasmids. Transformants were visualized by confocal microscopy. Bars, 5 μm. Lower panel: percentage of cells with no or small buds containing a round nucleus in wild-type and nem1Δ strains expressing GFP-Pus1. Three different transformants per strain were analyzed and for each one the number of cells counted was n=200. (D) Smp2 is evolutionarily conserved. Schematic representation of the primary structure of Smp2. The gray and black boxes indicate the highly conserved amino-terminal (N-lipin) and C-terminal (C-lipin) domains within Smp2. The percent sequence identity between the yeast domains and putative orthologues in various species is given.
